library(dplyr)
library(lubridate)
library(arrow)
library(readr)
library(climindi) # http://rfsaldanha.github.io/climindi/
library(zendown) # https://rfsaldanha.github.io/zendown/Introduction
Climatological normals are averages of climate variables observed for a given time – usually months – in a 30-year range. The normals are usually used as comparison benchmarks against recent or current conditions, being useful to recognize anomalies and to characterize global warming impacts.
The normals are usually computed with surface meteorological stations data, maintained by government meteorological institutions, and its distribution over the territory may be scarce or uneven, like in Brazil. Thus, the availability of climatological normals to different regions and administrative divisions like municipalities is difficult to obtain.
I propose here a method to compute monthly climatological normals for the 1961–1990 and monthly aggregated climate indicators from 1991 to 2023 for Brazilian municipalities using a climate reanalysis dataset.
In a hurry? Jump to the download section ;-)
Methods
A climatological normal can be computed with data from different sources, including remote sensing sensors and “area averages or points in gridded datasets” (WMO 2017). Some gridded climatological datasets are available for the Brazilian territory, including the ERA5-Land from Copernicus (Muñoz-Sabater et al. 2021) and the BR-DWGD dataset (Xavier et al. 2022), offering several climatological indicators for a long time range, and continuously updates.
Some research methods demands that climate data must be aggregated in the same spatial and temporal units of other data to be used in statistical models, being a fairly common procedure in epidemiology and economy studies. In order to approach this issue, spatial gridded data can be aggregated using zonal statistics (Saldanha et al. 2024).
I used the same methodology from the study above to create zonal statistics of climate indicators from the BR-DWGD project (Xavier et al. 2022). This data is available here, presenting climatological indicators from 1961 to March 2024 for all Brazilian municipalities. I propose here to compute the climatological normals and other climate aggregated indicators from this dataset of zonal statistics.
In order to compute these indicators, I created an R package named {climindi}. The package provides helper functions to compute climatological normals and other aggregated indicators in a tidy way.
Normal indicators
The {climindi} package computes the average, 10th and 90th percentile as climatological normals.
Aggregated indicators
The {climindi} package present functions to compute the following statistics for time-aggregated data: count of data points, average, median, standard deviation, standard error, maximum and minimum values, the 10th, 25th, 75th and 90th percentiles, and indicator-specific indicators, listed bellow.
- Precipitation
- Rain spells: count of rain spells occurrences, with 3 and 5 or more consecutive days with rain above the climatological normal average value
- Count of days with precipitation above 1mm, 5mm, 10mm, 50mm, and 100mm
- Count of sequences of 3 days, 5 days, 10 days, 15 days, 20 days, and 25 days or more without precipitation
- Maximum temperature
- Heat waves: Count of heat waves occurrences, with 3 and 5 or more consecutive days with maximum temperature above the climatological normal value plus 5 Celsius degrees
- Hot days: count of warm days, when the maximum temperature is above the normal 90th percentile
- Count of days with temperatures above or equal to 25, 30, 35, and 40 Celsius degrees
- Minimum temperature
- Cold spells: count of cold spells occurrences, with 3 and 5 or more consecutive days with minimum temperature bellow the climatological normal value minus 5 Celsius degrees
- Cold days: count of cold days, when the minimum temperature is bellow the normal 10th percentile
- Count of days with temperatures bellow or equal to 0, 5, 10, 15, and 20 Celsius degrees
- Relative humidity
- Count of dry spells occurrences, with 3 and 5 or more consecutive days with relative humidity bellow the climatological normal value minus 10 percent
- Count of wet spells occurrences, with 3 and 5 or more consecutive days with relative humidity above the climatological normal value plus 10 percent
- Count of dry days, when the relative humidity is bellow the normal 10th percentile
- Count of wet days, when the relative humidity is above the normal 90th percentile
- Count of days with relative humidity between 21% and 30% (Attention level)
- Count of days with relative humidity between 12% and 20% (Alert level)
- Count of days with relative humidity bellow 12% (Emergence level)
- Wind speed
- Count of sequences of 3 and 5 days or more with wind speed bellow the climatological average normal
- Count of sequences of 3 and 5 days or more with wind speed above the climatological average normal
- Evapotranspirations
- Count of sequences of 3 and 5 days or more with evapotranspirations bellow the climatological average normal
- Count of sequences of 3 and 5 days or more with evapotranspirations above the climatological average normal
- Solar radiation
- Count of sequences of 3 and 5 days or more with solar radiation bellow the climatological normal
- Count of sequences of 3 and 5 days or more with solar radiation above the climatological normal
Data source
The zonal statistics for the Brazilian municipalities computed with the BR-DWGD project is described here. We can use the {zendown} package to download the data files directly from from Zenodo.
Packages
To perform those computations, I needed to increase the envinroment variable R_MAX_VSIZE, as explained here.
Precipitation (mm)
Data
pr_data <- zen_file(13906834, "pr_3.2.3.parquet") |>
open_dataset() |>
filter(name == "pr_3.2.3_mean") |>
filter(date >= as.Date("1961-01-01")) |>
filter(date <= as.Date("2023-12-31")) |>
select(-name) |>
collect()Normal
pr_normal <- pr_data |>
# Identify month
mutate(month = month(date)) |>
# Group by id variable and month
group_by(code_muni, month) |>
# Compute normal
summarise_normal(
date_var = date,
value_var = value,
year_start = 1961,
year_end = 1990
) |>
# Ungroup
ungroup()Indicators
pr_indi <- pr_data |>
# Identify year
mutate(year = year(date)) |>
# Identify month
mutate(month = month(date)) |>
# Filter year
filter(year >= 1991) |>
# Group by id variable, year and month
group_by(code_muni, year, month) |>
# Compute precipitation indicators
summarise_precipitation(
value_var = value,
normals_df = pr_normal
) |>
# Ungroup
ungroup()Export
write_parquet(x = pr_normal, sink = "pr_normal.parquet")
write_csv2(x = pr_normal, file = "pr_normal.csv")
write_parquet(x = pr_indi, sink = "pr_indi.parquet")
write_csv2(x = pr_indi, file = "pr_indi.csv")Evapotranspiration (mm)
Data
eto_data <- zen_file(13906834, "ETo_3.2.3.parquet") |>
open_dataset() |>
filter(name == "ETo_3.2.3_mean") |>
filter(date >= as.Date("1961-01-01")) |>
filter(date <= as.Date("2023-12-31")) |>
select(-name) |>
collect()Normal
eto_normal <- eto_data |>
# Identify month
mutate(month = month(date)) |>
# Group by id variable and month
group_by(code_muni, month) |>
# Compute normal
summarise_normal(
date_var = date,
value_var = value,
year_start = 1961,
year_end = 1990
) |>
# Ungroup
ungroup()Indicators
eto_indi <- eto_data |>
# Identify year
mutate(year = year(date)) |>
# Identify month
mutate(month = month(date)) |>
# Filter year
filter(year >= 1991) |>
# Group by id variable, year and month
group_by(code_muni, year, month) |>
# Compute precipitation indicators
summarise_evapotrapiration(
value_var = value,
normals_df = eto_normal
) |>
# Ungroup
ungroup()Export
write_parquet(x = eto_normal, sink = "eto_normal.parquet")
write_csv2(x = eto_normal, file = "eto_normal.csv")
write_parquet(x = eto_indi, sink = "eto_indi.parquet")
write_csv2(x = eto_indi, file = "eto_indi.csv")Maximum temperature (°C)
Data
tmax_data <- zen_file(13906834, "Tmax_3.2.3.parquet") |>
open_dataset() |>
filter(name == "Tmax_3.2.3_mean") |>
filter(date >= as.Date("1961-01-01")) |>
filter(date <= as.Date("2023-12-31")) |>
select(-name) |>
collect()Normal
tmax_normal <- tmax_data |>
# Identify month
mutate(month = month(date)) |>
# Group by id variable and month
group_by(code_muni, month) |>
# Compute normal
summarise_normal(
date_var = date,
value_var = value,
year_start = 1961,
year_end = 1990
) |>
# Ungroup
ungroup()Indicators
tmax_indi <- tmax_data |>
# Identify year
mutate(year = year(date)) |>
# Identify month
mutate(month = month(date)) |>
# Filter year
filter(year >= 1991) |>
# Group by id variable, year and month
group_by(code_muni, year, month) |>
# Compute precipitation indicators
summarise_temp_max(
value_var = value,
normals_df = tmax_normal
) |>
# Ungroup
ungroup()Export
write_parquet(x = tmax_normal, sink = "tmax_normal.parquet")
write_csv2(x = tmax_normal, file = "tmax_normal.csv")
write_parquet(x = tmax_indi, sink = "tmax_indi.parquet")
write_csv2(x = tmax_indi, file = "tmax_indi.csv")Minimum temperature (°C)
Data
tmin_data <- zen_file(13906834, "Tmin_3.2.3.parquet") |>
open_dataset() |>
filter(name == "Tmin_3.2.3_mean") |>
filter(date >= as.Date("1961-01-01")) |>
filter(date <= as.Date("2023-12-31")) |>
select(-name) |>
collect()Normal
tmin_normal <- tmin_data |>
# Identify month
mutate(month = month(date)) |>
# Group by id variable and month
group_by(code_muni, month) |>
# Compute normal
summarise_normal(
date_var = date,
value_var = value,
year_start = 1961,
year_end = 1990
) |>
# Ungroup
ungroup()Indicators
tmin_indi <- tmin_data |>
# Identify year
mutate(year = year(date)) |>
# Identify month
mutate(month = month(date)) |>
# Filter year
filter(year >= 1991) |>
# Group by id variable, year and month
group_by(code_muni, year, month) |>
# Compute precipitation indicators
summarise_temp_min(
value_var = value,
normals_df = tmin_normal
) |>
# Ungroup
ungroup()Export
write_parquet(x = tmin_normal, sink = "tmin_normal.parquet")
write_csv2(x = tmin_normal, file = "tmin_normal.csv")
write_parquet(x = tmin_indi, sink = "tmin_indi.parquet")
write_csv2(x = tmin_indi, file = "tmin_indi.csv")Solar radiation (MJm-2)
Data
rs_data <- zen_file(13906834, "Rs_3.2.3.parquet") |>
open_dataset() |>
filter(name == "Rs_3.2.3_mean") |>
filter(date >= as.Date("1961-01-01")) |>
filter(date <= as.Date("2023-12-31")) |>
select(-name) |>
collect()Normal
rs_normal <- rs_data |>
# Identify month
mutate(month = month(date)) |>
# Group by id variable and month
group_by(code_muni, month) |>
# Compute normal
summarise_normal(
date_var = date,
value_var = value,
year_start = 1961,
year_end = 1990
) |>
# Ungroup
ungroup()Indicators
rs_indi <- rs_data |>
# Identify year
mutate(year = year(date)) |>
# Identify month
mutate(month = month(date)) |>
# Filter year
filter(year >= 1991) |>
# Group by id variable, year and month
group_by(code_muni, year, month) |>
# Compute precipitation indicators
summarise_solar_radiation(
value_var = value,
normals_df = rs_normal
) |>
# Ungroup
ungroup()Export
write_parquet(x = rs_normal, sink = "rs_normal.parquet")
write_csv2(x = rs_normal, file = "rs_normal.csv")
write_parquet(x = rs_indi, sink = "rs_indi.parquet")
write_csv2(x = rs_indi, file = "rs_indi.csv")Wind speed at 2m height (m/s)
Data
u2_data <- zen_file(13906834, "u2_3.2.3.parquet") |>
open_dataset() |>
filter(name == "u2_3.2.3_mean") |>
filter(date >= as.Date("1961-01-01")) |>
filter(date <= as.Date("2023-12-31")) |>
select(-name) |>
collect()Normal
u2_normal <- u2_data |>
# Identify month
mutate(month = month(date)) |>
# Group by id variable and month
group_by(code_muni, month) |>
# Compute normal
summarise_normal(
date_var = date,
value_var = value,
year_start = 1961,
year_end = 1990
) |>
# Ungroup
ungroup()Indicators
u2_indi <- u2_data |>
# Identify year
mutate(year = year(date)) |>
# Identify month
mutate(month = month(date)) |>
# Filter year
filter(year >= 1991) |>
# Group by id variable, year and month
group_by(code_muni, year, month) |>
# Compute precipitation indicators
summarise_windspeed(
value_var = value,
normals_df = u2_normal
) |>
# Ungroup
ungroup()Export
write_parquet(x = u2_normal, sink = "u2_normal.parquet")
write_csv2(x = u2_normal, file = "u2_normal.csv")
write_parquet(x = u2_indi, sink = "u2_indi.parquet")
write_csv2(x = u2_indi, file = "u2_indi.csv")Relative humidity (%)
Data
rh_data <- zen_file(13906834, "RH_3.2.3.parquet") |>
open_dataset() |>
filter(name == "RH_3.2.3_mean") |>
filter(date >= as.Date("1961-01-01")) |>
filter(date <= as.Date("2023-12-31")) |>
select(-name) |>
collect()Normal
rh_normal <- rh_data |>
# Identify month
mutate(month = month(date)) |>
# Group by id variable and month
group_by(code_muni, month) |>
# Compute normal
summarise_normal(
date_var = date,
value_var = value,
year_start = 1961,
year_end = 1990
) |>
# Ungroup
ungroup()Indicators
rh_indi <- rh_data |>
# Identify year
mutate(year = year(date)) |>
# Identify month
mutate(month = month(date)) |>
# Filter year
filter(year >= 1991) |>
# Group by id variable, year and month
group_by(code_muni, year, month) |>
# Compute precipitation indicators
summarise_rel_humidity(
value_var = value,
normals_df = rh_normal
) |>
# Ungroup
ungroup()Export
write_parquet(x = rh_normal, sink = "rh_normal.parquet")
write_csv2(x = rh_normal, file = "rh_normal.csv")
write_parquet(x = rh_indi, sink = "rh_indi.parquet")
write_csv2(x = rh_indi, file = "rh_indi.csv")Results and dataset download
The climatological normals and aggregated indicators of Brazilian municipalities can be downloaded from Zenodo on CSV and parquet formats. Click the link bellow to access and download the data.
You can also download the dataset directly from R, using the {zendown} package.
Let’s check some results.
Maximum temperature, Rio de Janeiro, RJ, 2023
Observed and normal
tmax_data <- zen_file(13906834, "Tmax_3.2.3.parquet") |>
open_dataset() |>
filter(name == "Tmax_3.2.3_mean") |>
filter(code_muni == 3304557) |>
filter(date >= as.Date("2023-01-01")) |>
filter(date <= as.Date("2023-12-31")) |>
select(-name) |>
collect()
tmax_normal <- zen_file(15519719, "tmax_normal.parquet") |>
open_dataset() |>
filter(code_muni == 3304557) |>
collect()library(ggplot2)
library(tidyr)
tmax_normal_exp <- tmax_normal |>
mutate(date = as_date(paste0("2023-", month, "-01"))) |>
group_by(month) %>%
expand(
date = seq.Date(
floor_date(date, unit = "month"),
ceiling_date(date, unit = "month") - days(1),
by = "day"
),
normal_mean,
normal_p10,
normal_p90
) |>
pivot_longer(cols = starts_with("normal_")) |>
mutate(name = substr(name, 8, 100))
ggplot() +
geom_line(data = tmax_data, aes(x = date, y = value)) +
geom_line(data = tmax_normal_exp, aes(x = date, y = value, color = name)) +
theme_bw() +
labs(
title = "Maximum temperature and climatological normal",
subtitle = "Rio de Janeiro, RJ",
color = "Normal (1961-1990)",
x = "Date",
y = "Celsius degrees"
) +
theme(legend.position = "bottom", legend.direction = "horizontal")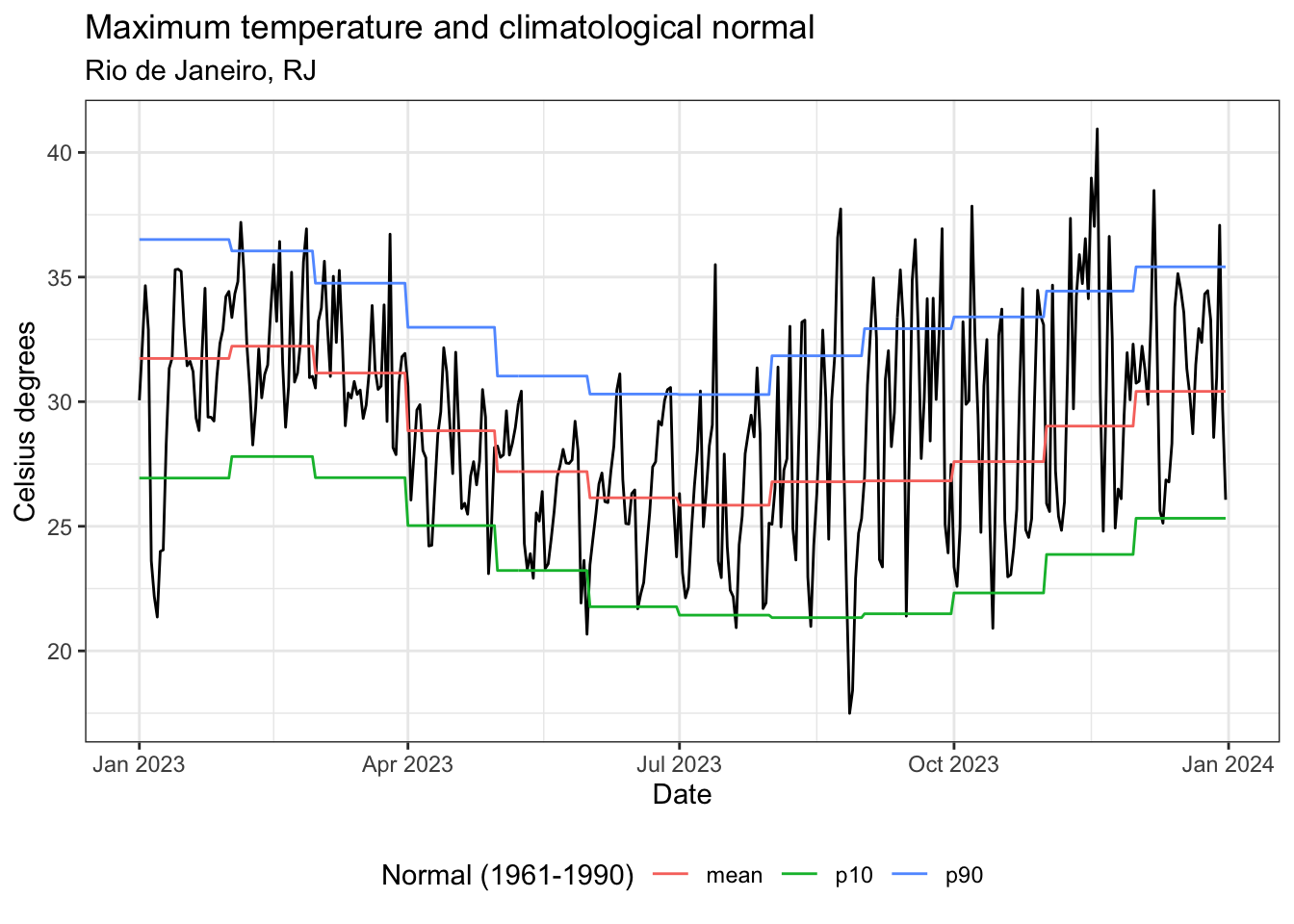
Indicators
library(gt)
zen_file(15519719, "tmax_indi.parquet") |>
open_dataset() |>
filter(code_muni == 3304557) |>
filter(year == 2023) |>
select(-code_muni, -year) |>
collect() |>
gt() |>
fmt_number(
columns = where(is.double),
decimals = 2,
use_seps = FALSE
)| month | count | normal_mean | normal_p10 | normal_p90 | mean | median | sd | se | max | min | p10 | p25 | p75 | p90 | heat_waves_3d | heat_waves_5d | hot_days | t_25 | t_30 | t_35 | t_40 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1.00 | 31 | 31.73 | 26.93 | 36.51 | 30.56 | 31.44 | 3.92 | 0.70 | 35.32 | 21.36 | 23.99 | 29.28 | 32.96 | 34.65 | 0 | 0 | 0 | 26 | 20 | 3 | 0 |
| 2.00 | 28 | 32.23 | 27.80 | 36.05 | 32.67 | 32.22 | 2.47 | 0.47 | 37.20 | 28.26 | 30.04 | 30.92 | 34.91 | 35.86 | 0 | 0 | 3 | 28 | 25 | 7 | 0 |
| 3.00 | 31 | 31.15 | 26.95 | 34.75 | 31.63 | 31.01 | 2.21 | 0.40 | 36.72 | 27.88 | 29.21 | 30.32 | 33.14 | 35.03 | 0 | 0 | 4 | 31 | 25 | 4 | 0 |
| 4.00 | 30 | 28.83 | 25.03 | 32.98 | 27.86 | 27.79 | 2.33 | 0.42 | 32.16 | 23.10 | 24.92 | 26.17 | 29.54 | 30.68 | 0 | 0 | 0 | 26 | 5 | 0 | 0 |
| 5.00 | 31 | 27.19 | 23.23 | 31.03 | 26.33 | 27.45 | 2.55 | 0.46 | 30.42 | 20.67 | 23.26 | 24.11 | 28.06 | 29.22 | 0 | 0 | 0 | 21 | 1 | 0 | 0 |
| 6.00 | 30 | 26.14 | 21.77 | 30.30 | 26.55 | 26.39 | 2.56 | 0.47 | 31.11 | 21.69 | 23.39 | 25.09 | 28.06 | 30.46 | 0 | 0 | 4 | 23 | 5 | 0 | 0 |
| 7.00 | 31 | 25.85 | 21.44 | 30.29 | 25.99 | 25.40 | 3.40 | 0.61 | 35.50 | 20.93 | 22.13 | 23.04 | 28.41 | 29.46 | 0 | 0 | 3 | 17 | 3 | 1 | 0 |
| 8.00 | 31 | 26.79 | 21.34 | 31.84 | 27.29 | 26.51 | 4.91 | 0.88 | 37.73 | 17.49 | 22.32 | 24.37 | 30.79 | 33.19 | 1 | 0 | 6 | 19 | 10 | 2 | 0 |
| 9.00 | 30 | 26.82 | 21.49 | 32.93 | 30.39 | 30.81 | 4.17 | 0.76 | 36.94 | 21.40 | 23.91 | 27.73 | 33.35 | 35.00 | 3 | 0 | 9 | 26 | 18 | 3 | 0 |
| 10.00 | 31 | 27.59 | 22.33 | 33.40 | 28.49 | 29.49 | 4.54 | 0.82 | 37.85 | 20.91 | 23.06 | 24.81 | 32.65 | 33.71 | 1 | 0 | 5 | 21 | 14 | 1 | 0 |
| 11.00 | 30 | 29.02 | 23.87 | 34.44 | 31.21 | 30.75 | 4.79 | 0.88 | 40.94 | 24.80 | 25.35 | 26.20 | 34.72 | 37.07 | 1 | 1 | 9 | 27 | 18 | 7 | 1 |
| 12.00 | 31 | 30.41 | 25.32 | 35.41 | 31.30 | 31.32 | 3.27 | 0.59 | 38.47 | 25.13 | 26.78 | 29.20 | 33.47 | 34.47 | 0 | 0 | 2 | 31 | 21 | 3 | 0 |
Session info
sessioninfo::session_info()─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.3.3 (2024-02-29)
os Ubuntu 24.04.2 LTS
system x86_64, linux-gnu
ui X11
language (EN)
collate en_US.UTF-8
ctype pt_BR.UTF-8
tz America/Sao_Paulo
date 2025-05-27
pandoc 3.1.3 @ /usr/bin/ (via rmarkdown)
quarto 1.6.42 @ /opt/quarto/bin/quarto
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
arrow * 20.0.0.2 2025-05-26 [1] CRAN (R 4.3.3)
assertthat 0.2.1 2019-03-21 [1] CRAN (R 4.3.3)
backports 1.5.0 2024-05-23 [1] CRAN (R 4.3.3)
bit 4.6.0 2025-03-06 [1] CRAN (R 4.3.3)
bit64 4.6.0-1 2025-01-16 [1] CRAN (R 4.3.3)
checkmate 2.3.2 2024-07-29 [1] CRAN (R 4.3.3)
cli 3.6.5 2025-04-23 [1] CRAN (R 4.3.3)
climindi * 0.1.0 2025-05-27 [1] Github (rfsaldanha/climindi@3acbdb9)
digest 0.6.37 2024-08-19 [1] CRAN (R 4.3.3)
dplyr * 1.1.4 2023-11-17 [1] CRAN (R 4.3.3)
evaluate 1.0.3 2025-01-10 [1] CRAN (R 4.3.3)
farver 2.1.2 2024-05-13 [1] CRAN (R 4.3.3)
fastmap 1.2.0 2024-05-15 [1] CRAN (R 4.3.3)
fs 1.6.6 2025-04-12 [1] CRAN (R 4.3.3)
generics 0.1.4 2025-05-09 [1] CRAN (R 4.3.3)
ggplot2 * 3.5.2 2025-04-09 [1] CRAN (R 4.3.3)
glue 1.8.0 2024-09-30 [1] CRAN (R 4.3.3)
gt * 1.0.0 2025-04-05 [1] CRAN (R 4.3.3)
gtable 0.3.6 2024-10-25 [1] CRAN (R 4.3.3)
hms 1.1.3 2023-03-21 [1] CRAN (R 4.3.3)
htmltools 0.5.8.1 2024-04-04 [1] CRAN (R 4.3.3)
htmlwidgets 1.6.4 2023-12-06 [1] CRAN (R 4.3.3)
jsonlite 2.0.0 2025-03-27 [1] CRAN (R 4.3.3)
knitr 1.50 2025-03-16 [1] CRAN (R 4.3.3)
labeling 0.4.3 2023-08-29 [1] CRAN (R 4.3.3)
lifecycle 1.0.4 2023-11-07 [1] CRAN (R 4.3.3)
lubridate * 1.9.4 2024-12-08 [1] CRAN (R 4.3.3)
magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.3.3)
pillar 1.10.2 2025-04-05 [1] CRAN (R 4.3.3)
pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.3.3)
purrr 1.0.4 2025-02-05 [1] CRAN (R 4.3.3)
R6 2.6.1 2025-02-15 [1] CRAN (R 4.3.3)
RColorBrewer 1.1-3 2022-04-03 [1] CRAN (R 4.3.3)
readr * 2.1.5 2024-01-10 [1] CRAN (R 4.3.3)
rlang 1.1.6 2025-04-11 [1] CRAN (R 4.3.3)
rmarkdown 2.29 2024-11-04 [1] CRAN (R 4.3.3)
sass 0.4.10 2025-04-11 [1] CRAN (R 4.3.3)
scales 1.4.0 2025-04-24 [1] CRAN (R 4.3.3)
sessioninfo 1.2.3 2025-02-05 [1] CRAN (R 4.3.3)
tibble 3.2.1 2023-03-20 [1] CRAN (R 4.3.3)
tidyr * 1.3.1 2024-01-24 [1] CRAN (R 4.3.3)
tidyselect 1.2.1 2024-03-11 [1] CRAN (R 4.3.3)
timechange 0.3.0 2024-01-18 [1] CRAN (R 4.3.3)
tzdb 0.5.0 2025-03-15 [1] CRAN (R 4.3.3)
vctrs 0.6.5 2023-12-01 [1] CRAN (R 4.3.3)
withr 3.0.2 2024-10-28 [1] CRAN (R 4.3.3)
xfun 0.52 2025-04-02 [1] CRAN (R 4.3.3)
xml2 1.3.8 2025-03-14 [1] CRAN (R 4.3.3)
yaml 2.3.10 2024-07-26 [1] CRAN (R 4.3.3)
zendown * 0.1.0 2024-05-21 [1] CRAN (R 4.3.3)
[1] /home/raphaelsaldanha/R/x86_64-pc-linux-gnu-library/4.3
[2] /usr/local/lib/R/site-library
[3] /usr/lib/R/site-library
[4] /usr/lib/R/library
* ── Packages attached to the search path.
──────────────────────────────────────────────────────────────────────────────