library(duckdb)
library(duckplyr)
library(glue)On ideal circumstances, any hospital admission would take place at the same city of residence of the patient. This facilitates the patient and family dislocation to the hospital, staying in a more familiar surrounding, and making the whole process less stressful. But, to a management point of view, this requires that all cities equal present a complete and extremely expensive set of hospital units, able to take care from simple diagnosis to complex organs transfusions
Thus, a national health system is usually organized under hierarchy and centralization principles. On this setting, smaller cities with less population have more simple resources and bigger cities have more complex resources at disposal. This administrative optimization comes with a cost: patients will likely need to travel to another city in order to receive a more complex health treatment, creating a flow of patients seeking for health care.
We will study here the flow of patients for hospital admissions using Brazilian datasets from its Universal Health System (SUS). This data is originally from the DataSUS, the informatics department of the Brazilian Health Ministry.
Dataset
We will use the Hospital Admissions data prepared by the PCDaS/Fiocruz with the original DataSUS data. This dataset is already cleaned, enriched and fully documented. I downloaded the CSV version, which contains 5,210 files, totaling 315.7 GB.
On these files, each row represents a hospital admission, and several variables of interest are available, including the patient’s municipality code of residence, the hospital’s code municipality and the date of the hospital admission.
It is a lot of data to process, making it very unpractical to just load the files into the computer memory.
The DuckDB database is very interesting for this case:
I will not need to create or have access to a database server, the DuckDB database is a just a file on your computer.
DuckDB have dedicated functions to parse and import CSV files straight to the database.
It is very fast for aggregate and other analytical functions that need to access all rows.
It is seamless integrated with R and
{dplyr}verbs.
Database creation
First, we need to call some packages and create the database and create an empty table on it.
- 1
- Create an empty database on the computer.
- 2
-
Create the
sihtable with a schema.
Now, we will populate the sih table with a loop.
years <- 2008:2023
for(y in years){
message(y)
query <- glue("INSERT INTO sih
SELECT res_codigo_adotado AS cod_res,
int_MUNCOD AS cod_int,
DT_INTER AS dt_inter, def_ident, def_car_int
FROM read_csv('/media/raphael/lacie/pcdas_sih/csv/*[y]*.csv',
delim = ',',
header = true,
dateformat = '%Y%m%d',
types = {'res_codigo_adotado': 'VARCHAR',
'int_MUNCOD': 'VARCHAR',
'dt_inter': 'DATE',
'def_ident': 'VARCHAR',
'def_car_int': 'VARCHAR'},
union_by_name = true
)", .open = "[", .close = "]")
dbExecute(con, query)
}
dbDisconnect(con)The query may seem a little complicated, but it is simple if we look at it by parts:
For each year
yfrom 2008 to 2023…Using the connection
con,INSERT INTOthe tablesihthe selected columns (changing its original names)FROMthe csv files. This is achieved with the DuckDB’s functionread_csv. Here, DuckDB will parse the CSV files contents that have the yearyon its name ([y].csv).
I tried to read all 5,210 files at once, but I received messages about “Too many files open”. This loop required fewer files to be open at the same time, being very effective.
One nice thing here is that we are reading into our table only the variables we want from the CSV files, reducing the database size and saving time.
After importing all CSV files, the database occupies 829MB on disk and the sih table have 187,735,977 rows.
Flow computation
Let’s compute the yearly flow of patients starting in 2008.
To compute the amount of patients that goes from a municipality to another on one year, we will do a grouping operation and observe the number of rows on each group.
1con <- dbConnect(duckdb(), dbdir = "../../flowsus/pcdas_sih.duckdb", read_only = FALSE)
2sih_tbl <- dplyr::tbl(con, "sih")
3res_geral <- sih_tbl |>
4 filter(def_ident == "Normal") |>
filter(year(dt_inter) >= 2008 & year(dt_inter) <= 2023) |>
5 mutate(year = year(dt_inter)) |>
summarise( #<56
6 weight = n(),
.by = c(year, cod_res, cod_int)
) |>
7 collect()
8dbDisconnect(con)- 1
- Connect with the database on read-only mode.
- 2
-
Create a virtual connection to the
sihtable. - 3
-
res_geralwill be the object that will receive the results. - 4
- Filter the hospital admissions, considering only the typical admissions (this excludes long-stays admissions, like mental healthcare) and filter hospital admissions that took place between 2008 and 2023.
- 5
- Create a new variable: the year of the hospital admission.
- 6
-
Summarize the table, by year and municipality of residence and destination, creating a
weightvariable that will receive the amount with rows. - 7
- Executes the query on the DuckDB database, returning a tibble ready-to-use.
- 8
- Disconnect the database.
The res_geral tibble present 1,395,512 rows.
head(res_geral)# A tibble: 6 × 4
year cod_res cod_int weight
<dbl> <chr> <chr> <dbl>
1 2008 292060 292250 100
2 2008 290323 290323 757
3 2008 292910 292250 5
4 2008 292260 291120 81
5 2008 291690 291760 36
6 2008 292340 292340 1327There are rows where the municipality of origin and destiny are the same. This represents the hospital admissions that took place at the same municipality of residence of the patient.
Discarding these loop-cases, the biggest observed flow occurred on 2021, when 28,375 hospital admissions occurred with patients from Jaboatão dos Guararapes, PE being admitted to hospitals from Recife, PE
res_geral |>
filter(cod_res != cod_int) |>
arrange(-weight) |>
head(10)# A tibble: 10 × 4
year cod_res cod_int weight
<dbl> <chr> <chr> <dbl>
1 2021 260790 261160 28375
2 2023 260790 261160 28329
3 2022 260790 261160 28142
4 2019 260790 261160 27268
5 2017 260790 261160 27222
6 2018 260790 261160 27025
7 2016 260790 261160 25254
8 2020 260790 261160 24134
9 2015 260790 261160 23970
10 2013 260790 261160 23718In total, 58,216,831 hospital admissions in Brazil occurred outside the patient’s residence municipality between 2008 and 2023.
res_geral |>
filter(cod_res != cod_int) |>
pull(weight) |>
sum()[1] 58216831And 124,835,812 hospital admissions in Brazil occurred at the same patient’s residence municipality between 2008 and 2023.
res_geral |>
filter(cod_res == cod_int) |>
pull(weight) |>
sum()[1] 124835812Map
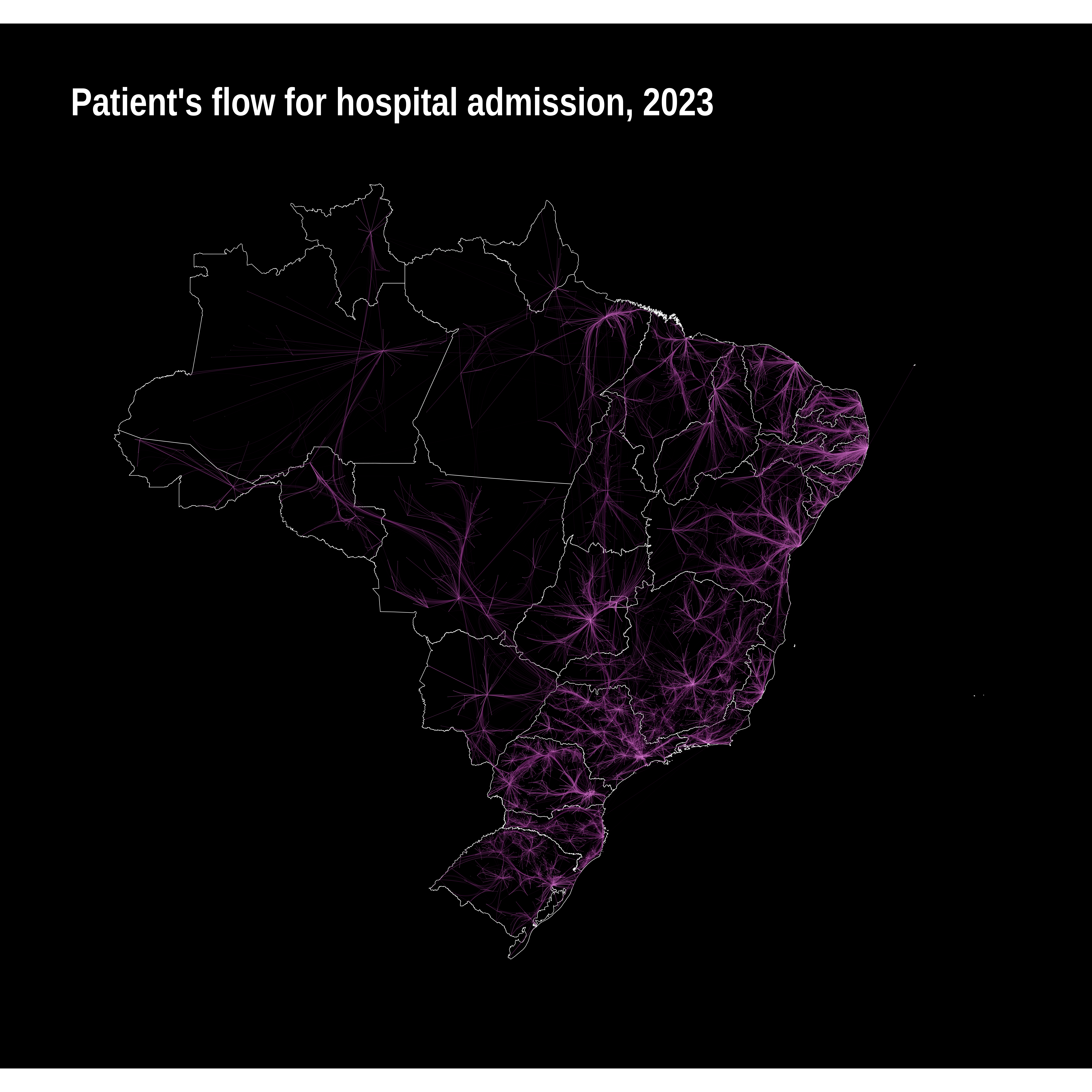
The flow of patients deserves a map! Let’s take a look at the last year available (2023).
We will need more packages.
library(dplyr)
library(geobr)
library(sf)
library(edgebundle)
library(igraph)
library(ggplot2)For the map, we will need the geographical coordinates of the municipalities and the state boundaries. The geobr package is very handy for this.
seats <- read_municipal_seat(showProgress = FALSE) |>
mutate(code_muni = substr(code_muni, 0, 6))Using year/date 2010states <- read_state(showProgress = FALSE)Using year/date 2010Prepare the municipal seats database.
seats <- seats |>
mutate(code_muni = substr(code_muni, 0, 6)) |>
mutate(longitude = st_coordinates(seats)[,1],
latitude = st_coordinates(seats)[,2]) |>
st_drop_geometry() |>
select(code_muni, longitude, latitude)
head(seats) code_muni longitude latitude
1 110001 -61.99982 -11.935540
2 110002 -63.03327 -9.908463
3 110003 -60.54431 -13.499763
4 110004 -61.44294 -11.433865
5 110005 -60.81843 -13.195033
6 110006 -60.55507 -13.130564And prepare the flow data for 2023. I will remove flows with less than 10 hospital admissions.
res_2023 <- res_geral |>
filter(weight >= 10) |>
filter(cod_res %in% seats$code_muni & cod_int %in% seats$code_muni) |>
filter(cod_res != cod_int) |>
filter(year == 2023) |>
select(2:4)
head(res_2023)# A tibble: 6 × 3
cod_res cod_int weight
<chr> <chr> <dbl>
1 291990 292400 91
2 293060 292720 30
3 290687 292720 88
4 291125 292720 16
5 292045 292840 44
6 292240 292870 565Now, we will create an igraph object to represent the network.
g_2023 <- graph_from_data_frame(d = res_2023, directed = TRUE, vertices = seats)And pairs of coordinates and vertices objects.
xy_2023 <- cbind(V(g_2023)$longitude, V(g_2023)$latitude)
verts_2023 <- data.frame(x = V(g_2023)$longitude, y = V(g_2023)$latitude)To plot the vertices, we will use a nice function from the edgebundle package. This may take some time to be computed.
pbundle_2023 <- edge_bundle_path(g_2023, xy_2023, max_distortion = 12, weight_fac = 2, segments = 50)
pbundle_2023 <- pbundle_2023 %>%
left_join(res_2023 |> mutate(id = row_number()) |> select(c(id, weight)), by=c('group' = 'id')) |>
st_as_sf(coords=c('x', 'y'), crs=4326) |>
group_by(group) |>
summarise(weight=mean(weight), do_union=FALSE) |>
arrange(weight) |>
st_cast("LINESTRING")Now, all needed objects are ready. Let’s plot it.
ggplot() +
geom_sf(
data = states,
col = "white", linewidth = 0.1, fill = NA
) +
geom_sf(
data = pbundle_2023, aes(group = group, linewidth = log(weight), color = log(weight), alpha = 0.5),
) +
scale_linewidth(range = c(0.001, .2)) +
scale_colour_gradient(low = "#3d0038", high = "#f993f1") +
geom_point(
data = verts_2023, aes(x, y),
stroke = 0,
col = "white", size = 0.1, alpha = 0.3
) +
labs(title = "Patient's flow for hospital admission, 2023") +
ggraph::theme_graph(background = "black") +
theme(plot.title = element_text(color = "white"), legend.position = "none")You can observe that some of the patient’s flow are its state of residence. On those cases, the patient is likely traveling to the state capital or to some big city within the state to receive healthcare. But the states size in Brazil varies a lot: the Amazonas state, by example, has an area bigger than Spain and France together. Also, other kind of flow occurs between states, typically from the interior or capital city from one state to the capital or regional center of the other state.
One important thing to observe is that some of those flows are expected, like the case where patients need to be admitted by hospitals with very specific capacities due to the patient’s disease and conditions.
The analysis of patient flows in a health system is very interesting and can guide health managers to better understand and organize the health system. I published with some colleagues on this theme some time ago ((Saldanha et al. 2019), (Xavier et al. 2019) and (Fonseca et al. 2022)).
Session info
sessioninfo::session_info()─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.4.0 (2024-04-24)
os Ubuntu 22.04.4 LTS
system x86_64, linux-gnu
ui X11
language en_US:en
collate en_US.UTF-8
ctype en_US.UTF-8
tz Europe/Paris
date 2024-06-12
pandoc 3.1.11 @ /usr/lib/rstudio/resources/app/bin/quarto/bin/tools/x86_64/ (via rmarkdown)
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
blob 1.2.4 2023-03-17 [1] CRAN (R 4.4.0)
cachem 1.1.0 2024-05-16 [1] CRAN (R 4.4.0)
class 7.3-22 2023-05-03 [4] CRAN (R 4.3.1)
classInt 0.4-10 2023-09-05 [1] CRAN (R 4.4.0)
cli 3.6.2 2023-12-11 [1] CRAN (R 4.4.0)
collections 0.3.7 2023-01-05 [1] CRAN (R 4.4.0)
colorspace 2.1-0 2023-01-23 [1] CRAN (R 4.4.0)
curl 5.2.1 2024-03-01 [1] CRAN (R 4.4.0)
data.table 1.15.4 2024-03-30 [1] CRAN (R 4.4.0)
DBI * 1.2.3 2024-06-02 [1] CRAN (R 4.4.0)
dbplyr 2.5.0 2024-03-19 [1] CRAN (R 4.4.0)
digest 0.6.35 2024-03-11 [1] CRAN (R 4.4.0)
dplyr * 1.1.4 2023-11-17 [1] CRAN (R 4.4.0)
duckdb * 0.10.2 2024-05-01 [1] CRAN (R 4.4.0)
duckplyr * 0.4.0 2024-05-21 [1] CRAN (R 4.4.0)
e1071 1.7-14 2023-12-06 [1] CRAN (R 4.4.0)
edgebundle * 0.4.2 2023-12-16 [1] CRAN (R 4.4.0)
evaluate 0.24.0 2024-06-10 [1] CRAN (R 4.4.0)
fansi 1.0.6 2023-12-08 [1] CRAN (R 4.4.0)
farver 2.1.2 2024-05-13 [1] CRAN (R 4.4.0)
fastmap 1.2.0 2024-05-15 [1] CRAN (R 4.4.0)
generics 0.1.3 2022-07-05 [1] CRAN (R 4.4.0)
geobr * 1.9.0 2024-04-18 [1] CRAN (R 4.4.0)
ggforce 0.4.2 2024-02-19 [1] CRAN (R 4.4.0)
ggplot2 * 3.5.1 2024-04-23 [1] CRAN (R 4.4.0)
ggraph 2.2.1 2024-03-07 [1] CRAN (R 4.4.0)
ggrepel 0.9.5 2024-01-10 [1] CRAN (R 4.4.0)
glue * 1.7.0 2024-01-09 [1] CRAN (R 4.4.0)
graphlayouts 1.1.1 2024-03-09 [1] CRAN (R 4.4.0)
gridExtra 2.3 2017-09-09 [1] CRAN (R 4.4.0)
gtable 0.3.5 2024-04-22 [1] CRAN (R 4.4.0)
htmltools 0.5.8.1 2024-04-04 [1] CRAN (R 4.4.0)
htmlwidgets 1.6.4 2023-12-06 [1] CRAN (R 4.4.0)
httr 1.4.7 2023-08-15 [1] CRAN (R 4.4.0)
igraph * 2.0.3 2024-03-13 [1] CRAN (R 4.4.0)
jsonlite 1.8.8 2023-12-04 [1] CRAN (R 4.4.0)
KernSmooth 2.23-24 2024-05-17 [4] CRAN (R 4.4.0)
knitr 1.47 2024-05-29 [1] CRAN (R 4.4.0)
labeling 0.4.3 2023-08-29 [1] CRAN (R 4.4.0)
lattice 0.22-5 2023-10-24 [4] CRAN (R 4.3.1)
lifecycle 1.0.4 2023-11-07 [1] CRAN (R 4.4.0)
magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.4.0)
MASS 7.3-60 2023-05-04 [4] CRAN (R 4.3.1)
Matrix 1.6-5 2024-01-11 [4] CRAN (R 4.3.3)
memoise 2.0.1 2021-11-26 [1] CRAN (R 4.4.0)
munsell 0.5.1 2024-04-01 [1] CRAN (R 4.4.0)
pillar 1.9.0 2023-03-22 [1] CRAN (R 4.4.0)
pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.4.0)
png 0.1-8 2022-11-29 [1] CRAN (R 4.4.0)
polyclip 1.10-6 2023-09-27 [1] CRAN (R 4.4.0)
proxy 0.4-27 2022-06-09 [1] CRAN (R 4.4.0)
purrr 1.0.2 2023-08-10 [1] CRAN (R 4.4.0)
R6 2.5.1 2021-08-19 [1] CRAN (R 4.4.0)
Rcpp 1.0.12 2024-01-09 [1] CRAN (R 4.4.0)
reticulate 1.37.0 2024-05-21 [1] CRAN (R 4.4.0)
rlang 1.1.4 2024-06-04 [1] CRAN (R 4.4.0)
rmarkdown 2.27 2024-05-17 [1] CRAN (R 4.4.0)
rstudioapi 0.16.0 2024-03-24 [1] CRAN (R 4.4.0)
scales 1.3.0 2023-11-28 [1] CRAN (R 4.4.0)
sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.4.0)
sf * 1.0-16 2024-03-24 [1] CRAN (R 4.4.0)
tibble 3.2.1 2023-03-20 [1] CRAN (R 4.4.0)
tidygraph 1.3.1 2024-01-30 [1] CRAN (R 4.4.0)
tidyr 1.3.1 2024-01-24 [1] CRAN (R 4.4.0)
tidyselect 1.2.1 2024-03-11 [1] CRAN (R 4.4.0)
tweenr 2.0.3 2024-02-26 [1] CRAN (R 4.4.0)
units 0.8-5 2023-11-28 [1] CRAN (R 4.4.0)
utf8 1.2.4 2023-10-22 [1] CRAN (R 4.4.0)
vctrs 0.6.5 2023-12-01 [1] CRAN (R 4.4.0)
viridis 0.6.5 2024-01-29 [1] CRAN (R 4.4.0)
viridisLite 0.4.2 2023-05-02 [1] CRAN (R 4.4.0)
withr 3.0.0 2024-01-16 [1] CRAN (R 4.4.0)
xfun 0.44 2024-05-15 [1] CRAN (R 4.4.0)
yaml 2.3.8 2023-12-11 [1] CRAN (R 4.4.0)
[1] /home/raphael/R/x86_64-pc-linux-gnu-library/4.4
[2] /usr/local/lib/R/site-library
[3] /usr/lib/R/site-library
[4] /usr/lib/R/library
──────────────────────────────────────────────────────────────────────────────