gm_br_uf <- readRDS(file = "gm_br_uf.rds")
DT::datatable(gm_br_uf)Introduction
Rates allow the comparison between the number of counts between multiple classes with different population sizes. For example, 10 disease cases that occur in 100 population region have a different proportional importance than 10 cases in a 1,000 population region.
In epidemiology, those overall rates may be compared if both populations are similar. If the populations have different constitutions, the comparison between overall rates may be misleading as the disease may affect the age groups differently.
Available packages
Some R packages, such as epitools and epiR, allow the computation of crude and adjusted rates. Those packages use as input, vectors or matrices containing the events count and population per age group.
The downside of them is that you need to use the rates function manually repeatedly times if you are going to compute rates for several regions.
tidyrates package
I created the tidyrates to compute direct and indirect adjusted rates for several regions, years, and other keys that you might use. Internally, the package wraps the epitools functions and applies them to each group.
Example: General mortality rates in Brazilian states, from 2010 to 2021
Mortality data
The dataset bellow presents the total number of deaths by any cause (general mortality) for the Brazilian states per age group, from 2010 to 2021. I collected the data from the PCDaS platform.
Population data
To compute rates, we will need the state population size per age group for each year. The brpop package present estimates for this. We just need to do some adaptations.
- 1
-
Get population table from
brpoppackage. - 2
- Filter the desired years
- 3
- Rename the population variable
- 4
- Remove population totals, keeping only the population per age group
- 5
- Change year variable from numeric to character.
DT::datatable(pop_uf)Reference population
To compute standardized rates, we need a reference population. We may use the SEER reference population (which is a corrected version from the WHO reference population) that is present in the tidyrates package. We need to use the same age group labels and also merge the population from 80 years or more to make it compatible with our data.
seer2 <- tidyrates::seer_std_pop %>%
mutate(age_group = case_when(
age_group == "0-4" ~ "From 0 to 4 years",
age_group == "5-9" ~ "From 5 to 9 years",
age_group == "10-14" ~ "From 10 to 14 years",
age_group == "15-19" ~ "From 15 to 19 years",
age_group == "20-24" ~ "From 20 to 24 years",
age_group == "25-29" ~ "From 25 to 29 years",
age_group == "30-34" ~ "From 30 to 34 years",
age_group == "35-39" ~ "From 35 to 39 years",
age_group == "40-44" ~ "From 40 to 44 years",
age_group == "45-49" ~ "From 45 to 49 years",
age_group == "50-54" ~ "From 50 to 54 years",
age_group == "55-59" ~ "From 55 to 59 years",
age_group == "60-64" ~ "From 60 to 64 years",
age_group == "65-69" ~ "From 65 to 69 years",
age_group == "70-74" ~ "From 70 to 74 years",
age_group == "75-79" ~ "From 75 to 79 years",
age_group == "80-84" ~ "From 80 years or more",
age_group == "85-89" ~ "From 80 years or more",
age_group == "90-94" ~ "From 80 years or more",
age_group == "95-99" ~ "From 80 years or more",
age_group == "100+" ~ "From 80 years or more",
.default = age_group
)) %>%
group_by(age_group) %>%
summarise(population = sum(population)) %>%
ungroup()DT::datatable(seer2)Crude and direct adjusted rates
Now, we can join the mortality dataset with the population dataset by uf, year, and age groups.
gm_pop <- left_join(gm_br_uf, pop_uf, by = c("uf", "year", "age_group")) %>%
pivot_longer(cols = c("events", "population"))
DT::datatable(gm_pop)With the data ready, we can compute the direct adjusted rates with the tidyrates package for all UFs and years.
library(tidyrates)
rates <- rate_adj_direct(.data = gm_pop, .std = seer2, .keys = c("year", "uf"))
DT::datatable(rates)Plot
To make a plot, let’s do some modifications and pivot the data.
library(ggplot2)
library(geofacet)
rates_for_plot <- rates %>%
mutate(year = as.numeric(year)) %>%
pivot_longer(cols = c("crude.rate", "adj.rate", "lci", "uci")) %>%
filter(name %in% c("crude.rate", "adj.rate")) %>%
mutate(value = value * 100000)
ci_for_plot <- rates %>%
mutate(year = as.numeric(year)) %>%
select(year, uf, lci, uci) %>%
mutate(
lci = lci * 100000,
uci = uci * 100000
)
br_grid <- br_states_grid1 %>%
mutate(code_num = case_when(
code == "RO" ~ 11,
code == "AC" ~ 12,
code == "AM" ~ 13,
code == "RR" ~ 14,
code == "PA" ~ 15,
code == "AP" ~ 16,
code == "TO" ~ 17,
code == "MA" ~ 21,
code == "PI" ~ 22,
code == "CE" ~ 23,
code == "RN" ~ 24,
code == "PB" ~ 25,
code == "PE" ~ 26,
code == "AL" ~ 27,
code == "SE" ~ 28,
code == "BA" ~ 29,
code == "MG" ~ 31,
code == "ES" ~ 32,
code == "RJ" ~ 33,
code == "SP" ~ 35,
code == "PR" ~ 41,
code == "SC" ~ 42,
code == "RS" ~ 43,
code == "MS" ~ 50,
code == "MT" ~ 51,
code == "GO" ~ 52,
code == "DF" ~ 53,
))ggplot(data = rates_for_plot) +
geom_line(aes(x = year, y = value, color = name)) +
geom_ribbon(data = ci_for_plot, aes(x = year, ymin = lci, ymax = uci), alpha=0.3, fill = "lightpink", color = "pink") +
facet_geo(~uf, grid = br_grid, label = "name") +
theme_bw() +
theme(legend.position = "bottom", legend.direction = "horizontal") +
labs(color = "Rate", x = "Year", y = "Rate per 100,000 inhab.")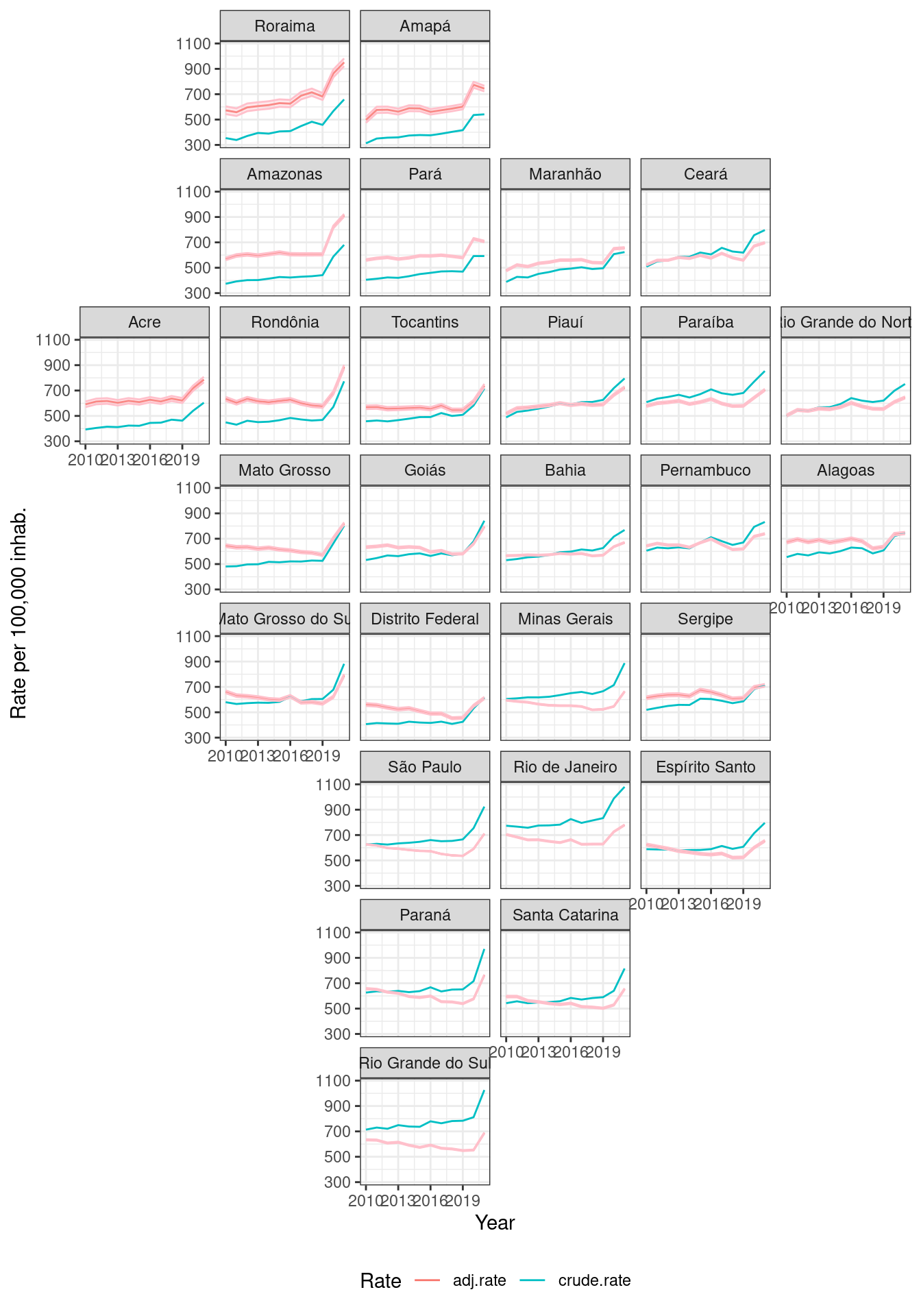
Session info
sessionInfo()R version 4.3.2 (2023-10-31)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 22.04.3 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.10.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.10.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_CA.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_CA.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_CA.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_CA.UTF-8 LC_IDENTIFICATION=C
time zone: Europe/Paris
tzcode source: system (glibc)
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] geofacet_0.2.0 ggplot2_3.4.4 tidyrates_0.0.1 brpop_0.3.0
[5] tidyr_1.3.0 dplyr_1.1.3
loaded via a namespace (and not attached):
[1] gtable_0.3.4 xfun_0.41 bslib_0.5.1
[4] htmlwidgets_1.6.2 ggrepel_0.9.4 processx_3.8.2
[7] lattice_0.22-5 RApiSerialize_0.1.2 callr_3.7.3
[10] vctrs_0.6.4 tools_4.3.2 crosstalk_1.2.0
[13] ps_1.7.5 generics_0.1.3 parallel_4.3.2
[16] tibble_3.2.1 proxy_0.4-27 fansi_1.0.5
[19] pkgconfig_2.0.3 KernSmooth_2.23-22 checkmate_2.3.0
[22] RcppParallel_5.1.7 lifecycle_1.0.3 farver_2.1.1
[25] compiler_4.3.2 multidplyr_0.1.3 munsell_0.5.0
[28] qs_0.25.5 codetools_0.2-19 imguR_1.0.3
[31] htmltools_0.5.7 class_7.3-22 sass_0.4.7
[34] yaml_2.3.7 pillar_1.9.0 jquerylib_0.1.4
[37] ellipsis_0.3.2 epitools_0.5-10.1 classInt_0.4-10
[40] DT_0.30 cachem_1.0.8 parallelly_1.36.0
[43] tidyselect_1.2.0 digest_0.6.33 future_1.33.0
[46] sf_1.0-14 purrr_1.0.2 listenv_0.9.0
[49] labeling_0.4.3 forcats_1.0.0 rnaturalearth_0.3.4
[52] geogrid_0.1.2 fastmap_1.1.1 grid_4.3.2
[55] colorspace_2.1-0 cli_3.6.1 magrittr_2.0.3
[58] utf8_1.2.4 e1071_1.7-13 withr_2.5.2
[61] scales_1.2.1 backports_1.4.1 sp_2.1-1
[64] httr_1.4.7 rmarkdown_2.25 jpeg_0.1-10
[67] globals_0.16.2 gridExtra_2.3 png_0.1-8
[70] stringfish_0.15.8 evaluate_0.23 knitr_1.45
[73] rlang_1.1.2 Rcpp_1.0.11 glue_1.6.2
[76] DBI_1.1.3 rstudioapi_0.15.0 jsonlite_1.8.7
[79] R6_2.5.1 units_0.8-4