library(tidyverse)
library(lubridate)
library(arrow)
library(timetk)
library(dtwclust)
library(kableExtra)
library(tictoc)
source("../functions.R")Raw cases
This notebook aims to cluster the Brazilian municipalities considering dengue raw cases time-series similarities.
Packages
Load data
Load the bundled data.
tdengue <- open_dataset(sources = data_dir("bundled_data/tdengue.parquet")) %>%
select(mun, date, cases = cases_raw) %>%
collect()
dim(tdengue)[1] 340179 3Prepare data
The chunk bellow formats the dataset for tsclust use.
tdengue <- tdengue %>%
# Prepare time series
arrange(mun) %>%
pivot_wider(names_from = mun, values_from = cases) %>%
select(-date) %>%
t() %>%
# Convert object
tslist()length(tdengue)[1] 679Clustering
Sequence of k groups to be used.
k_seq <- 2:10DTW (basic)
tic()
clust_dtw <- tsclust(
series = tdengue,
type = "partitional",
k = k_seq,
distance = "dtw_basic",
seed = 13
)
toc()46.292 sec elapsednames(clust_dtw) <- paste0("k_", k_seq)
res_cvi <- sapply(clust_dtw, cvi, type = "internal") %>%
t() %>%
as_tibble(rownames = "k") %>%
arrange(-Sil)
res_cvi %>%
gt::gt()| k | Sil | SF | CH | DB | DBstar | D | COP |
|---|---|---|---|---|---|---|---|
| k_2 | 0.563306808 | 0 | 243.13362 | 2.777906 | 2.777906 | 4.033050e-03 | 0.81493978 |
| k_3 | 0.500486555 | 0 | 179.80850 | 2.589921 | 8.236031 | 3.463657e-03 | 0.06040454 |
| k_5 | 0.234604708 | 0 | 102.69761 | 2.868848 | 23.661427 | 7.946198e-04 | 0.05528690 |
| k_4 | 0.221410311 | 0 | 122.82411 | 2.535349 | 24.240538 | 7.813649e-04 | 0.07362652 |
| k_8 | 0.047380416 | 0 | 71.53960 | 3.385318 | 105.686817 | 3.503914e-04 | 0.06330307 |
| k_7 | 0.018931701 | 0 | 66.01800 | 3.523758 | 87.921436 | 2.131907e-04 | 0.06658659 |
| k_10 | -0.005982317 | 0 | 52.35818 | 3.393992 | 207.433508 | 1.469383e-04 | 0.03558253 |
| k_9 | -0.011726942 | 0 | 64.19814 | 3.436507 | 244.767080 | 1.527283e-04 | 0.02848551 |
| k_6 | -0.101115607 | 0 | 73.21168 | 4.166345 | 409.352091 | 1.633515e-05 | 0.06955750 |
sel_clust <- clust_dtw[[res_cvi[[1,1]]]]
plot(sel_clust)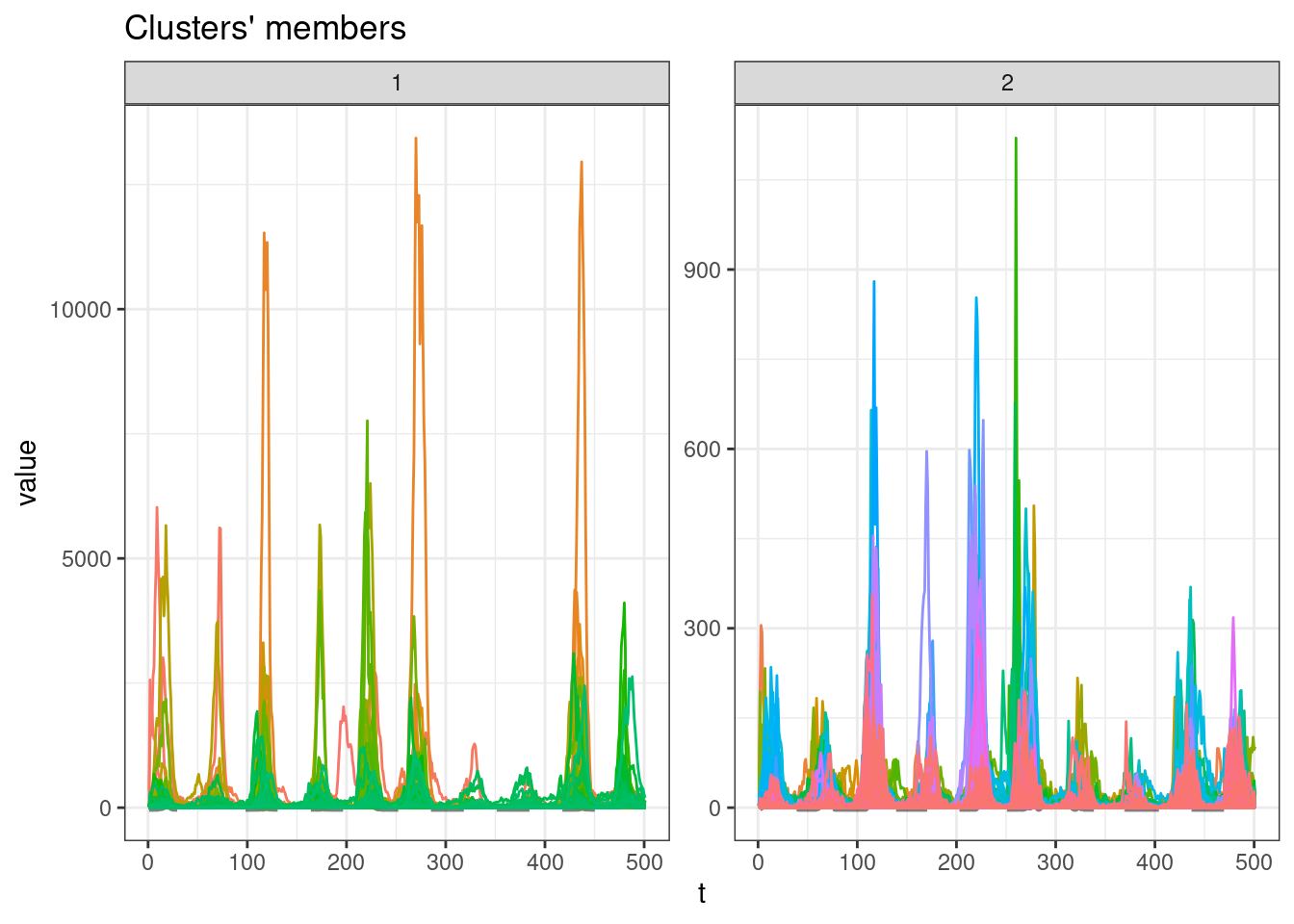
plot(sel_clust, type = "centroids", lty = 1)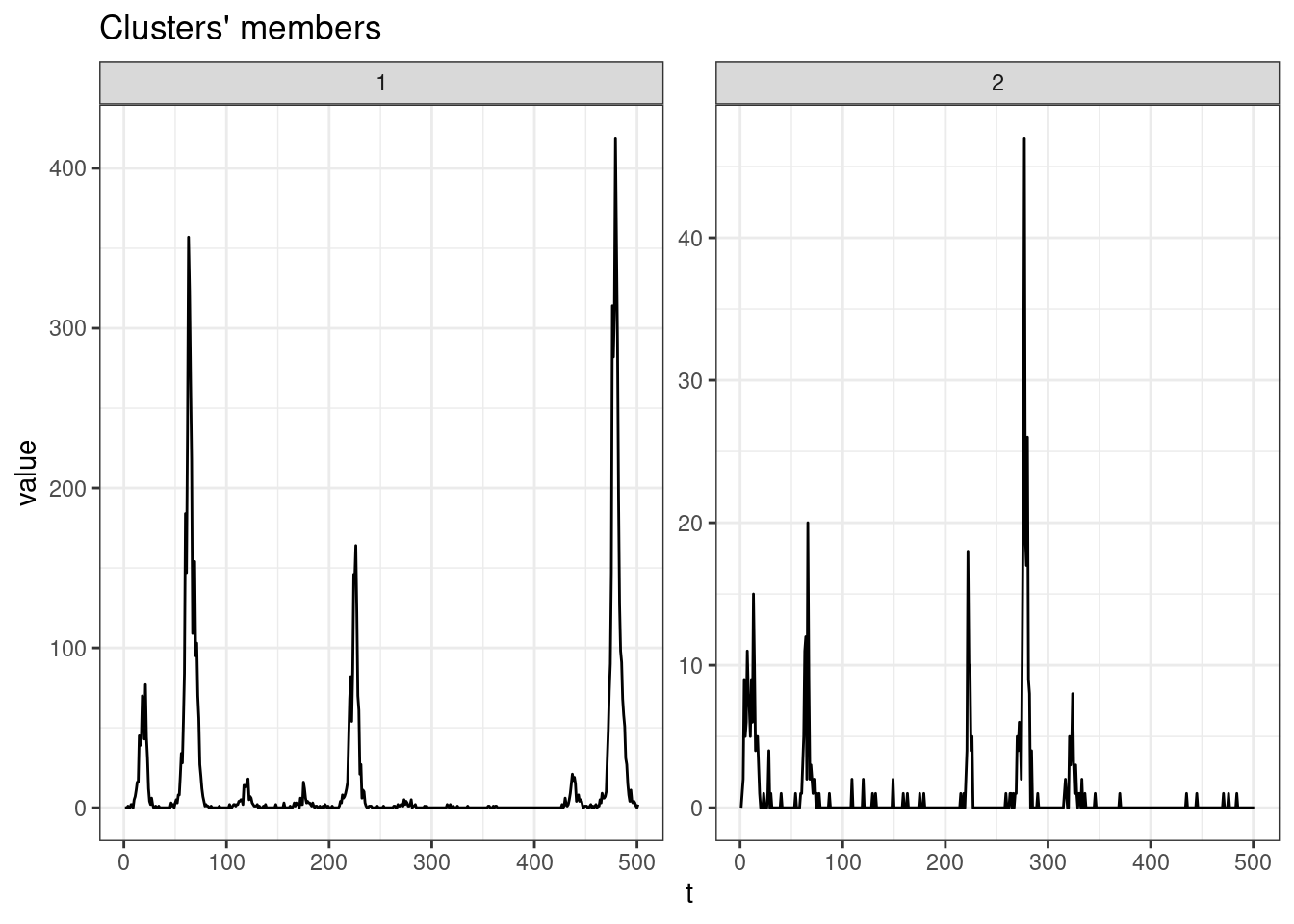
table(sel_clust@cluster)
1 2
187 492 Soft-DTW
tic()
clust_sdtw <- tsclust(
series = tdengue,
type = "partitional",
k = k_seq,
distance = "sdtw",
seed = 13
)
toc()231.662 sec elapsednames(clust_sdtw) <- paste0("k_", k_seq)
res_cvi <- sapply(clust_sdtw, cvi, type = "internal") %>%
t() %>%
as_tibble(rownames = "k") %>%
arrange(-Sil)
res_cvi %>%
gt::gt()| k | Sil | SF | CH | DB | DBstar | D | COP |
|---|---|---|---|---|---|---|---|
| k_2 | 0.9747238 | 0 | 610.6681 | 0.7127038 | 7.127038e-01 | 6.394018e-03 | 0.021962268 |
| k_3 | 0.8785473 | 0 | 339.7721 | 1.3846248 | 2.184955e+01 | 6.047589e-05 | 0.012882348 |
| k_4 | 0.7831408 | 0 | 367.3748 | 1.0620911 | 8.628989e+01 | 3.151199e-05 | 0.006517102 |
| k_5 | 0.5616851 | 0 | 287.9459 | 1.6410825 | 6.279292e+02 | 3.508382e-06 | 0.006502140 |
| k_6 | 0.4127518 | 0 | 232.3673 | 1.8248896 | 1.900511e+03 | 1.029877e-06 | 0.006496556 |
| k_9 | 0.3492449 | 0 | 161.4531 | 2.1597151 | 6.103722e+03 | 5.179334e-07 | 0.006423601 |
| k_8 | 0.3130712 | 0 | 165.0773 | 2.5475305 | 5.893653e+03 | 6.826592e-07 | 0.006458184 |
| k_7 | 0.1779085 | 0 | 192.9691 | 2.0300445 | 3.657527e+04 | 1.456487e-07 | 0.006500423 |
| k_10 | 0.1726132 | 0 | 130.6724 | 1.9249465 | 1.215560e+05 | 7.778736e-08 | 0.006444721 |
sel_clust <- clust_sdtw[[res_cvi[[1,1]]]]
plot(sel_clust)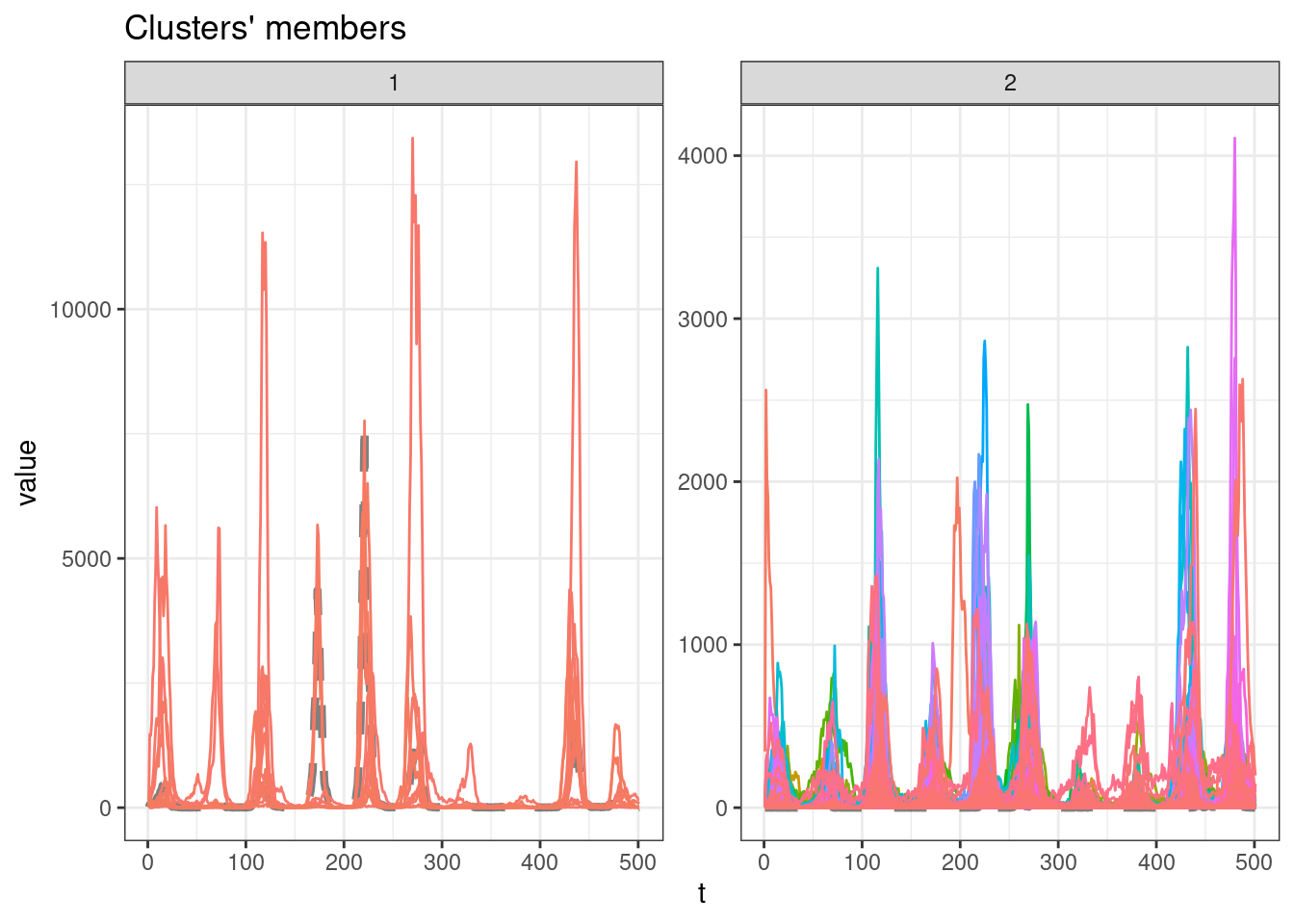
plot(sel_clust, type = "centroids", lty = 1)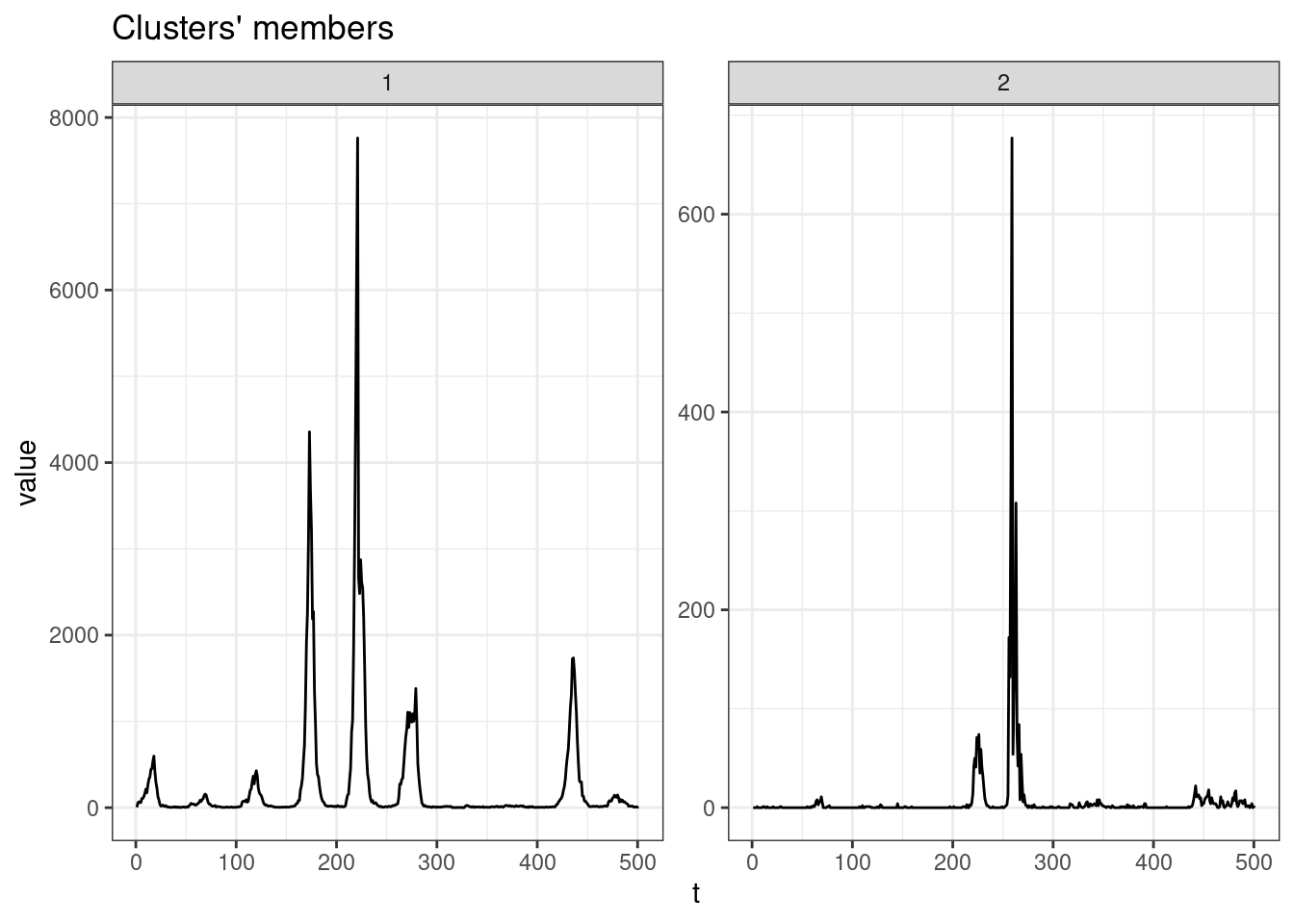
table(sel_clust@cluster)
1 2
11 668 SBD
tic()
clust_sbd <- tsclust(
series = tdengue,
type = "partitional",
k = k_seq,
distance = "sbd",
seed = 13
)
toc()0.945 sec elapsednames(clust_sbd) <- paste0("k_", k_seq)
res_cvi <- sapply(clust_sbd, cvi, type = "internal") %>%
t() %>%
as_tibble(rownames = "k") %>%
arrange(-Sil)
res_cvi %>%
gt::gt()| k | Sil | SF | CH | DB | DBstar | D | COP |
|---|---|---|---|---|---|---|---|
| k_2 | 0.19232458 | 0.43352764 | 105.89969 | 3.495106 | 3.495106 | 0.04758953 | 0.3749125 |
| k_6 | 0.16139544 | 0.14186007 | 42.69029 | 3.539215 | 3.955084 | 0.05571189 | 0.3521767 |
| k_5 | 0.13008017 | 0.20138965 | 70.46372 | 2.731617 | 3.777455 | 0.05647432 | 0.3400697 |
| k_10 | 0.09211874 | 0.04440823 | 49.64036 | 2.799473 | 4.241962 | 0.05073587 | 0.3159727 |
| k_3 | 0.08978010 | 0.31394588 | 37.06136 | 6.461761 | 7.845973 | 0.02136961 | 0.3667538 |
| k_9 | 0.05631414 | 0.06024602 | 55.09867 | 2.170914 | 3.178624 | 0.01656789 | 0.3179895 |
| k_4 | 0.04322383 | 0.28795370 | 96.18451 | 4.350614 | 4.857858 | 0.01715419 | 0.3501107 |
| k_8 | 0.03766029 | 0.08963975 | 48.19501 | 3.360877 | 4.869244 | 0.01731394 | 0.3257173 |
| k_7 | 0.03553741 | 0.11301210 | 70.16402 | 2.846658 | 4.211674 | 0.02132175 | 0.3251809 |
sel_clust <- clust_sbd[[res_cvi[[1,1]]]]
plot(sel_clust)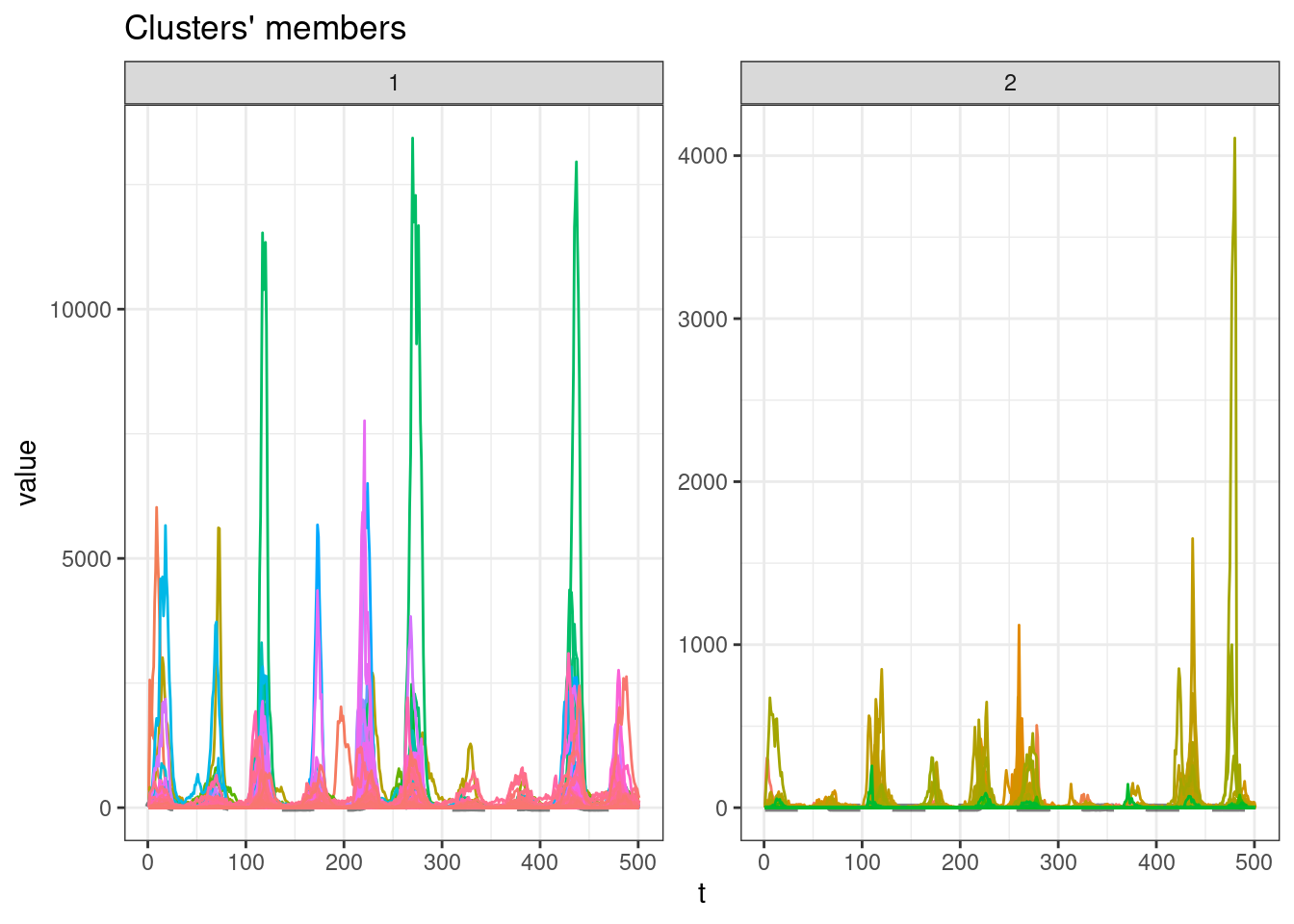
plot(sel_clust, type = "centroids", lty = 1)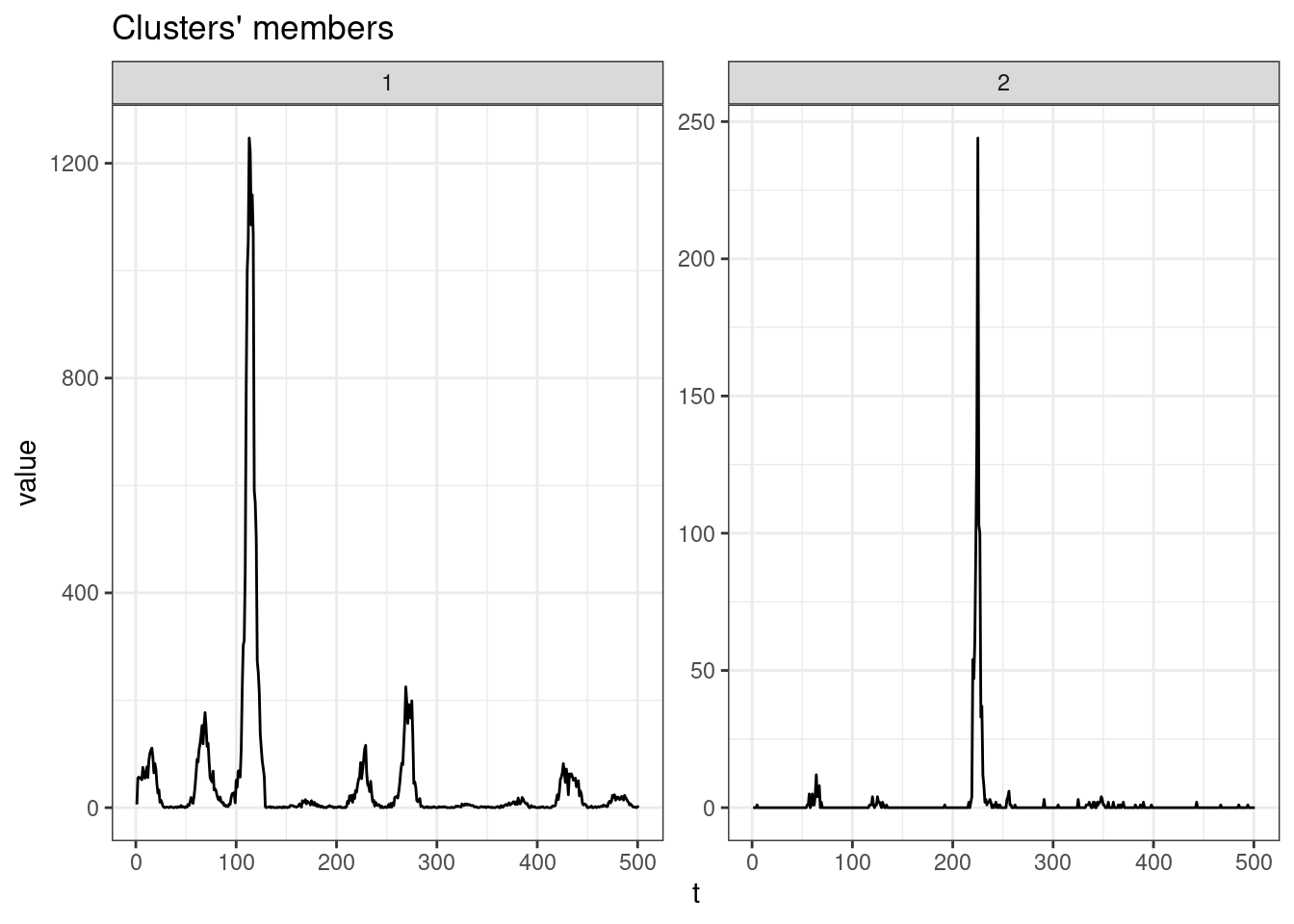
table(sel_clust@cluster)
1 2
511 168 GAK
tic()
clust_gak <- tsclust(
series = tdengue,
type = "partitional",
k = k_seq,
distance = "gak",
seed = 13
)
toc()271.303 sec elapsednames(clust_gak) <- paste0("k_", k_seq)
res_cvi <- sapply(clust_gak, cvi, type = "internal") %>%
t() %>%
as_tibble(rownames = "k") %>%
arrange(-Sil)
res_cvi %>%
gt::gt()| k | Sil | SF | CH | DB | DBstar | D | COP |
|---|---|---|---|---|---|---|---|
| k_2 | 0.91204464 | 0.6121344 | 70.160389 | 11.397435 | 11.39743 | 1.070841e-03 | 0.024308301 |
| k_3 | 0.89063056 | 0.6025345 | 35.827115 | 12.214961 | 21.33743 | 8.882340e-04 | 0.017392584 |
| k_8 | 0.56673089 | 0.5761049 | 9.540570 | 36.984617 | 772.68089 | 2.073418e-05 | 0.009652511 |
| k_4 | 0.51422407 | 0.6090642 | 37.881634 | 9.134131 | 109.09898 | 2.369289e-05 | 0.023596187 |
| k_7 | 0.34615432 | 0.5804793 | 10.401018 | 24.242925 | 872.28716 | 2.770548e-06 | 0.012348716 |
| k_5 | 0.32153320 | 0.5967512 | 18.167789 | 15.597700 | 963.89750 | 2.566534e-06 | 0.017167921 |
| k_10 | 0.30902233 | 0.5821542 | 9.743672 | 20.658947 | 759.39685 | 3.587207e-07 | 0.011523184 |
| k_9 | 0.19782535 | 0.5908109 | 11.123215 | 20.521645 | 2053.75182 | 3.399318e-07 | 0.019667996 |
| k_6 | 0.09332721 | 0.5976834 | 24.094674 | 9.259298 | 1869.00352 | 3.435826e-07 | 0.016792580 |
sel_clust <- clust_gak[[res_cvi[[1,1]]]]
plot(sel_clust)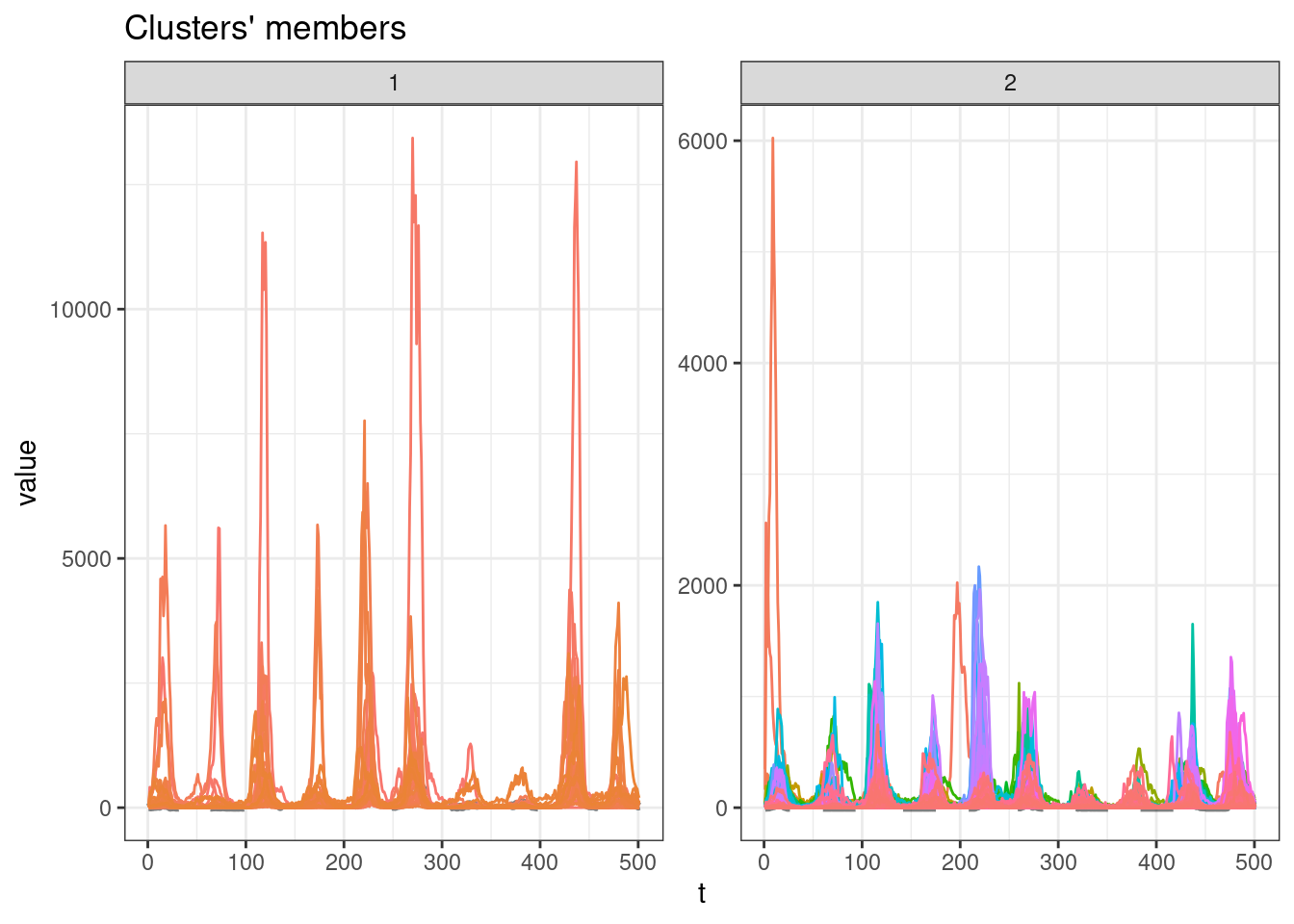
plot(sel_clust, type = "centroids", lty = 1)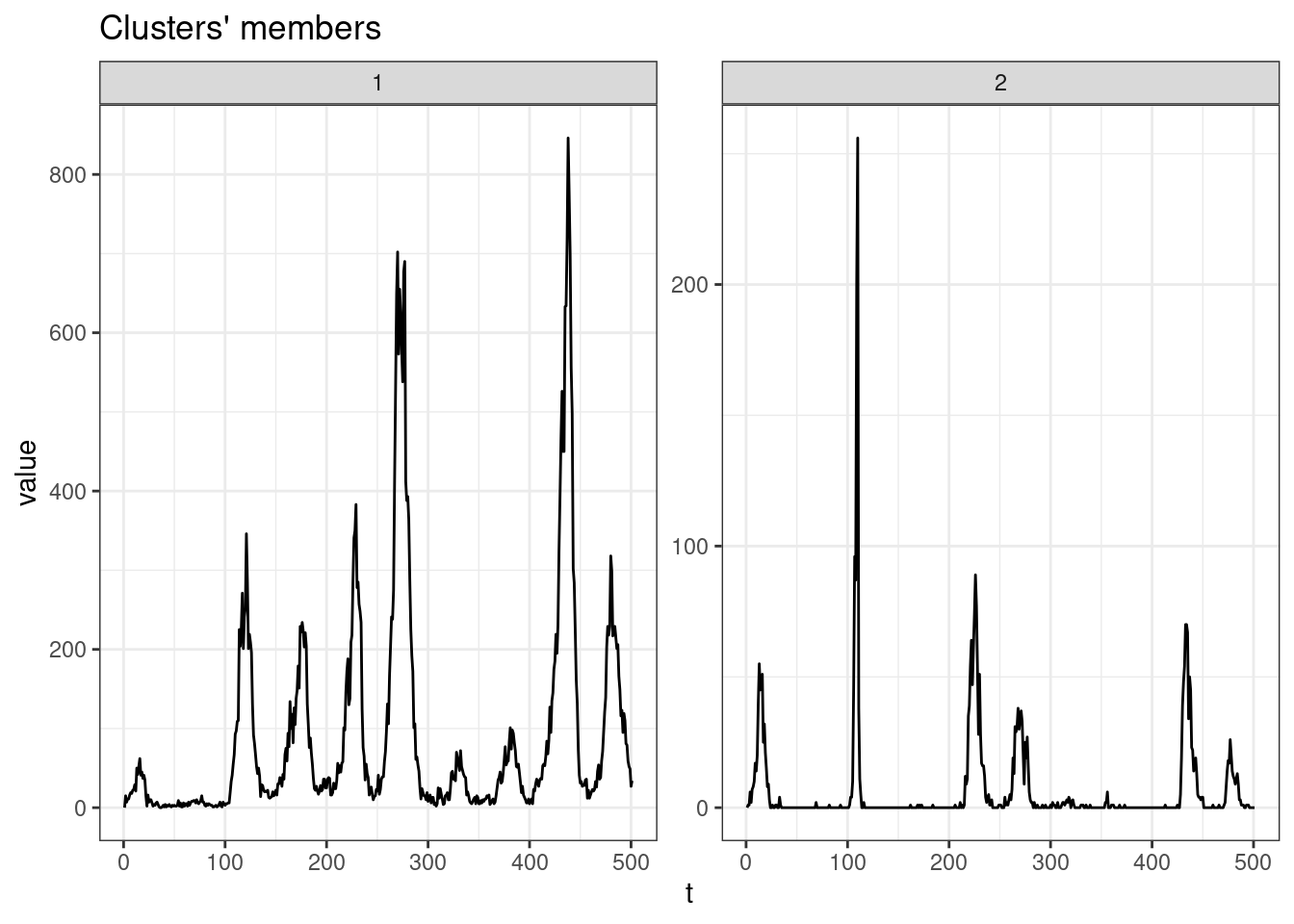
table(sel_clust@cluster)
1 2
32 647 Session info
sessionInfo()R version 4.3.2 (2023-10-31)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 22.04.3 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.10.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.10.0
Random number generation:
RNG: L'Ecuyer-CMRG
Normal: Inversion
Sample: Rejection
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_CA.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_CA.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_CA.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_CA.UTF-8 LC_IDENTIFICATION=C
time zone: Europe/Paris
tzcode source: system (glibc)
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] tictoc_1.2 kableExtra_1.3.4 dtwclust_5.5.12 dtw_1.23-1
[5] proxy_0.4-27 timetk_2.9.0 arrow_13.0.0.1 lubridate_1.9.3
[9] forcats_1.0.0 stringr_1.5.0 dplyr_1.1.3 purrr_1.0.2
[13] readr_2.1.4 tidyr_1.3.0 tibble_3.2.1 ggplot2_3.4.4
[17] tidyverse_2.0.0
loaded via a namespace (and not attached):
[1] rlang_1.1.2 magrittr_2.0.3 clue_0.3-65
[4] furrr_0.3.1 flexclust_1.4-1 compiler_4.3.2
[7] systemfonts_1.0.5 vctrs_0.6.4 reshape2_1.4.4
[10] rvest_1.0.3 lhs_1.1.6 tune_1.1.2
[13] pkgconfig_2.0.3 fastmap_1.1.1 ellipsis_0.3.2
[16] labeling_0.4.3 utf8_1.2.4 promises_1.2.1
[19] rmarkdown_2.25 prodlim_2023.08.28 tzdb_0.4.0
[22] bit_4.0.5 xfun_0.41 modeltools_0.2-23
[25] jsonlite_1.8.7 recipes_1.0.8 later_1.3.1
[28] parallel_4.3.2 cluster_2.1.4 R6_2.5.1
[31] stringi_1.7.12 rsample_1.2.0 parallelly_1.36.0
[34] rpart_4.1.21 Rcpp_1.0.11 assertthat_0.2.1
[37] dials_1.2.0 iterators_1.0.14 knitr_1.45
[40] future.apply_1.11.0 zoo_1.8-12 httpuv_1.6.12
[43] Matrix_1.6-1.1 splines_4.3.2 nnet_7.3-19
[46] timechange_0.2.0 tidyselect_1.2.0 rstudioapi_0.15.0
[49] yaml_2.3.7 timeDate_4022.108 codetools_0.2-19
[52] listenv_0.9.0 lattice_0.22-5 plyr_1.8.9
[55] shiny_1.7.5.1 withr_2.5.2 evaluate_0.23
[58] future_1.33.0 survival_3.5-7 RcppParallel_5.1.7
[61] xml2_1.3.5 xts_0.13.1 pillar_1.9.0
[64] foreach_1.5.2 stats4_4.3.2 shinyjs_2.1.0
[67] generics_0.1.3 hms_1.1.3 munsell_0.5.0
[70] scales_1.2.1 xtable_1.8-4 globals_0.16.2
[73] class_7.3-22 glue_1.6.2 tools_4.3.2
[76] data.table_1.14.8 RSpectra_0.16-1 webshot_0.5.5
[79] gower_1.0.1 grid_4.3.2 yardstick_1.2.0
[82] ipred_0.9-14 colorspace_2.1-0 cli_3.6.1
[85] DiceDesign_1.9 workflows_1.1.3 parsnip_1.1.1
[88] fansi_1.0.5 viridisLite_0.4.2 gt_0.10.0
[91] svglite_2.1.2 lava_1.7.3 gtable_0.3.4
[94] GPfit_1.0-8 sass_0.4.7 digest_0.6.33
[97] ggrepel_0.9.4 farver_2.1.1 htmlwidgets_1.6.2
[100] htmltools_0.5.7 lifecycle_1.0.4 httr_1.4.7
[103] hardhat_1.3.0 mime_0.12 bit64_4.0.5
[106] MASS_7.3-60