library(tidyverse)
library(lubridate)
library(arrow)
library(timetk)
library(dtwclust)
library(kableExtra)
library(tictoc)
source("../functions.R")Yearly time series
This notebook aims to cluster the Brazilian municipalities considering yearly dengue cases time-series similarities.
Packages
Load data
Load the bundled data.
dengue <- open_dataset(sources = data_dir("bundled_data/tdengue.parquet")) %>%
select(mun, date, cases = cases_raw) %>%
collect()
dim(dengue)[1] 340179 3Prepare data
The chunk bellow executes various steps to prepare the data for clustering.
tdengue <- dengue %>%
# Remove dates
filter(date >= as.Date("2011-01-01") & date < as.Date("2020-01-01")) %>%
# Create year variable
mutate(year = year(date)) %>%
# Create week number
mutate(week = epiweek(date)) %>%
# Summarise per year and isoweek
group_by(mun, year, week) %>%
summarise(cases = sum(cases, na.rm = TRUE)) %>%
# Scale cases
mutate(cases = scale(cases)) %>%
ungroup() %>%
# Isolate municipality and year
mutate(mun = paste0(mun, "_", year)) %>%
select(-year, week) %>%
# Arrange data
arrange(mun, week) %>%
# Prepare time series of municipalities by year
mutate(mun = paste0("m_", mun)) %>%
pivot_wider(names_from = mun, values_from = cases) %>%
select(-week) %>%
# Use zero value for years withot week 53
mutate(across(everything(), ~replace_na(.x, 0))) %>%
# Transpose as matrix
t() %>%
# Convert object
tslist()`summarise()` has grouped output by 'mun', 'year'. You can override using the
`.groups` argument.length(tdengue)[1] 6111Clustering
Sequence of k groups to be used.
k_seq <- 3:10Soft-DTW method
tic()
clust <- tsclust(
series = tdengue,
type = "partitional",
k = k_seq,
distance = "sdtw",
seed = 12
)
toc()186.312 sec elapsedCluster Validity Indices (CVI)
names(clust) <- paste0("k_", k_seq)
res_cvi <- sapply(clust, cvi, type = "internal") %>%
t() %>%
as_tibble(rownames = "k") %>%
arrange(-Sil)
res_cvi %>%
kbl() %>%
kable_styling()| k | Sil | SF | CH | DB | DBstar | D | COP |
|---|---|---|---|---|---|---|---|
| k_7 | 0.3042978 | 0 | 1498.9650 | 1.643061 | 2.034144 | -0.0090704 | 0.1154715 |
| k_5 | 0.3009208 | 0 | 1828.2539 | 1.510974 | 2.111730 | -0.0046130 | 0.2050362 |
| k_3 | 0.3000788 | 0 | 2147.2455 | 1.141345 | 1.363098 | -0.0037204 | 0.2592231 |
| k_6 | 0.2851064 | 0 | 1873.0643 | 1.799201 | 2.168677 | -0.0044977 | 0.1324393 |
| k_8 | 0.2586174 | 0 | 1636.1817 | 2.149713 | 3.552313 | -0.0065691 | 0.1022790 |
| k_9 | 0.2489799 | 0 | 958.2712 | 3.232229 | 3.805038 | -0.0057935 | 0.1829836 |
| k_10 | 0.2028457 | 0 | 1416.5421 | 2.552099 | 4.802588 | -0.0078132 | 0.0904757 |
| k_4 | 0.1600588 | 0 | 1613.0683 | 2.871479 | 4.394944 | -0.0054940 | 0.2373967 |
Cluster with higher Silhouette statistic
sel_clust <- clust[[res_cvi[[1,1]]]]plot(sel_clust)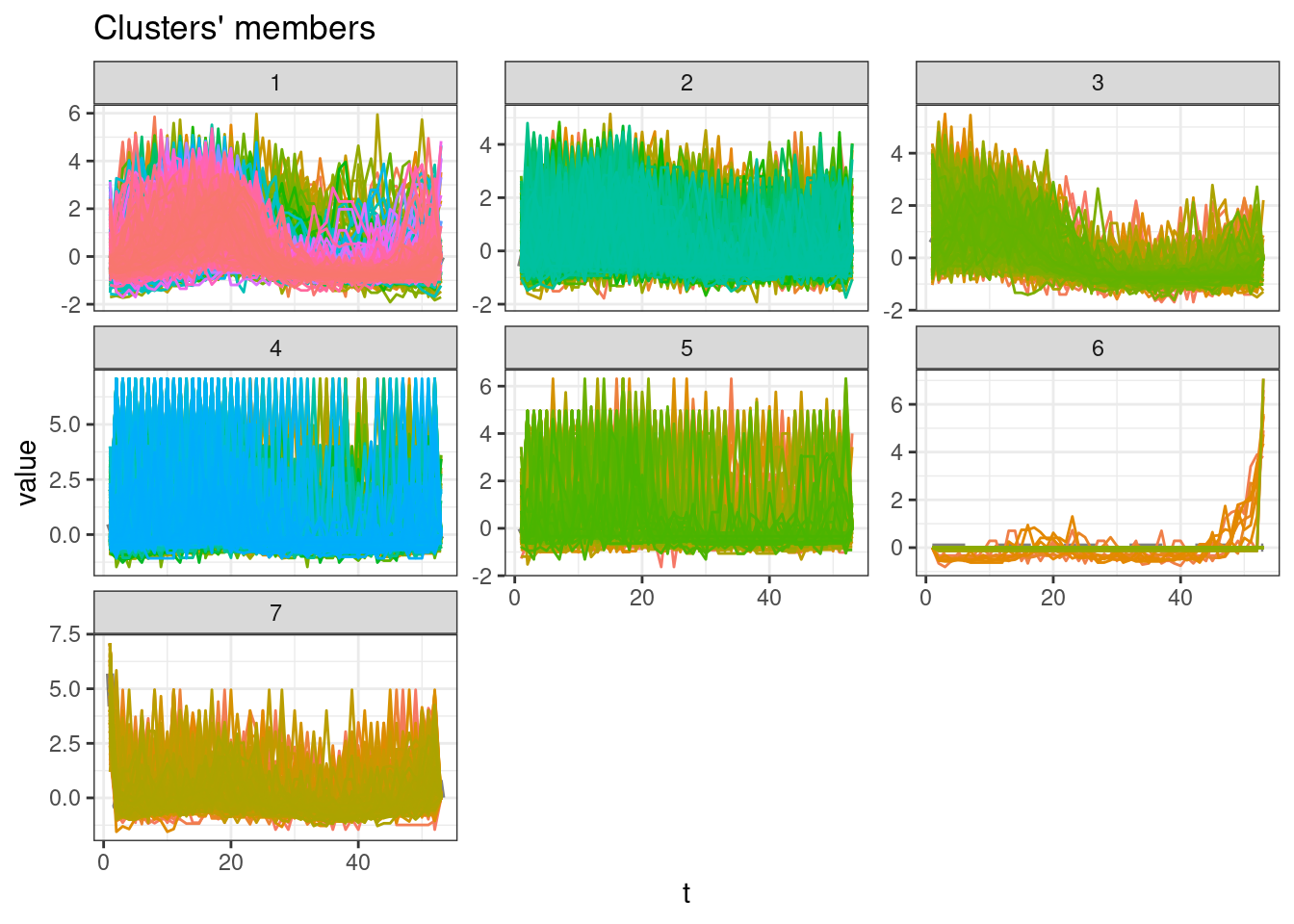
ggsave(filename = "cluster_mun_year.pdf")Saving 7 x 5 in imageplot(sel_clust, type = "centroids", lty = 1)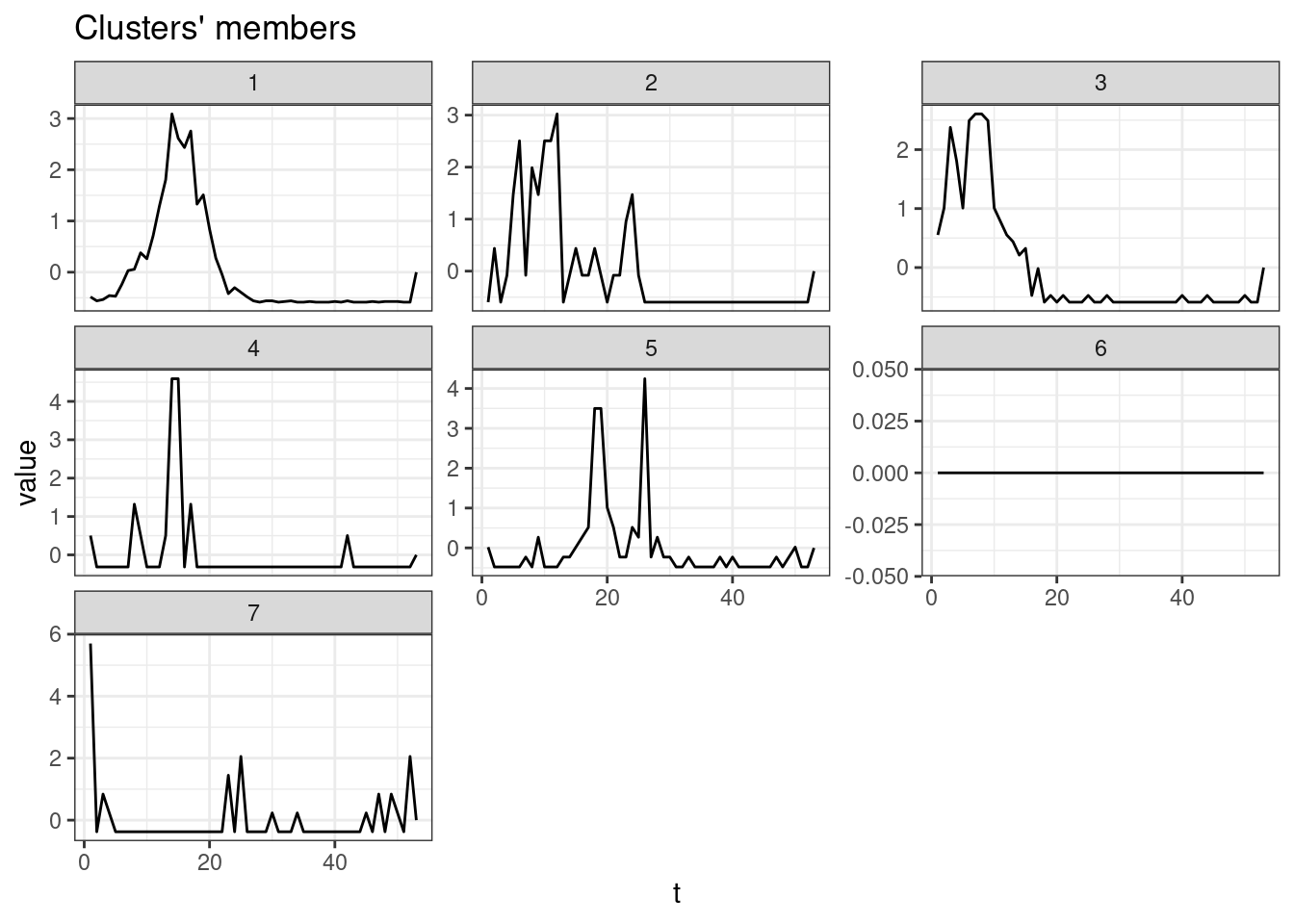
ggsave(filename = "cluster_mun_year_centr.pdf")Saving 7 x 5 in imageCluster sizes
table(sel_clust@cluster)
1 2 3 4 5 6 7
2008 895 551 1227 587 466 377 Session info
sessionInfo()R version 4.3.2 (2023-10-31)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 22.04.3 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.10.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.10.0
Random number generation:
RNG: L'Ecuyer-CMRG
Normal: Inversion
Sample: Rejection
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_CA.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_CA.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_CA.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_CA.UTF-8 LC_IDENTIFICATION=C
time zone: Europe/Paris
tzcode source: system (glibc)
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] tictoc_1.2 kableExtra_1.3.4 dtwclust_5.5.12 dtw_1.23-1
[5] proxy_0.4-27 timetk_2.9.0 arrow_13.0.0.1 lubridate_1.9.3
[9] forcats_1.0.0 stringr_1.5.0 dplyr_1.1.3 purrr_1.0.2
[13] readr_2.1.4 tidyr_1.3.0 tibble_3.2.1 ggplot2_3.4.4
[17] tidyverse_2.0.0
loaded via a namespace (and not attached):
[1] rlang_1.1.2 magrittr_2.0.3 clue_0.3-65
[4] furrr_0.3.1 flexclust_1.4-1 compiler_4.3.2
[7] systemfonts_1.0.5 vctrs_0.6.4 reshape2_1.4.4
[10] rvest_1.0.3 lhs_1.1.6 tune_1.1.2
[13] pkgconfig_2.0.3 fastmap_1.1.1 ellipsis_0.3.2
[16] labeling_0.4.3 utf8_1.2.4 promises_1.2.1
[19] rmarkdown_2.25 prodlim_2023.08.28 tzdb_0.4.0
[22] ragg_1.2.6 bit_4.0.5 xfun_0.41
[25] modeltools_0.2-23 jsonlite_1.8.7 recipes_1.0.8
[28] highr_0.10 later_1.3.1 parallel_4.3.2
[31] cluster_2.1.4 R6_2.5.1 stringi_1.7.12
[34] rsample_1.2.0 parallelly_1.36.0 rpart_4.1.21
[37] Rcpp_1.0.11 assertthat_0.2.1 dials_1.2.0
[40] iterators_1.0.14 knitr_1.45 future.apply_1.11.0
[43] zoo_1.8-12 httpuv_1.6.12 Matrix_1.6-1.1
[46] splines_4.3.2 nnet_7.3-19 timechange_0.2.0
[49] tidyselect_1.2.0 rstudioapi_0.15.0 yaml_2.3.7
[52] timeDate_4022.108 codetools_0.2-19 listenv_0.9.0
[55] lattice_0.22-5 plyr_1.8.9 shiny_1.7.5.1
[58] withr_2.5.2 evaluate_0.23 future_1.33.0
[61] survival_3.5-7 RcppParallel_5.1.7 xml2_1.3.5
[64] xts_0.13.1 pillar_1.9.0 foreach_1.5.2
[67] stats4_4.3.2 shinyjs_2.1.0 generics_0.1.3
[70] hms_1.1.3 munsell_0.5.0 scales_1.2.1
[73] xtable_1.8-4 globals_0.16.2 class_7.3-22
[76] glue_1.6.2 tools_4.3.2 data.table_1.14.8
[79] RSpectra_0.16-1 webshot_0.5.5 gower_1.0.1
[82] grid_4.3.2 yardstick_1.2.0 ipred_0.9-14
[85] colorspace_2.1-0 cli_3.6.1 DiceDesign_1.9
[88] textshaping_0.3.7 workflows_1.1.3 parsnip_1.1.1
[91] fansi_1.0.5 viridisLite_0.4.2 svglite_2.1.2
[94] lava_1.7.3 gtable_0.3.4 GPfit_1.0-8
[97] digest_0.6.33 ggrepel_0.9.4 farver_2.1.1
[100] htmlwidgets_1.6.2 htmltools_0.5.7 lifecycle_1.0.4
[103] httr_1.4.7 hardhat_1.3.0 mime_0.12
[106] bit64_4.0.5 MASS_7.3-60