library(tidyverse)
library(tidymodels)
library(arrow)
library(timetk)
library(rpart.plot)
library(vip)Classification task
This notebook models the relationship between dengue cases and weather variables using a classification of dengue cases as outbreak level (anomaly) or base level.
Packages
Dengue data
Subset and aggregate
Rio de Janeiro, RJ, aggregated by month.
dengue_rj <- open_dataset("../dengue-data/parquet_aggregated/dengue_md.parquet") %>%
filter(mun == 330455) %>%
collect() %>%
summarise_by_time(.date_var = date, .by = "month", freq = sum(freq, na.rm = TRUE))plot_time_series(.data = dengue_rj, .date_var = date, .value = freq, .smooth = FALSE, .title = "Dengue, absolute number of cases")Classify
plot_anomaly_diagnostics(.data = dengue_rj, .date_var = date, .value = freq, .alpha = 0.10, .max_anomalies = 1, .legend_show = FALSE)frequency = 12 observations per 1 yeartrend = 60 observations per 5 yearsdengue_rj_anom <- tk_anomaly_diagnostics(.data = dengue_rj, .date_var = date, .value = freq, .alpha = 0.10, .max_anomalies = 1) %>%
select(date, anomaly) %>%
mutate(anomaly = as.factor(anomaly))frequency = 12 observations per 1 yeartrend = 60 observations per 5 yearsdengue_rj <- inner_join(dengue_rj, dengue_rj_anom) Joining with `by = join_by(date)`prop.table(table(dengue_rj$anomaly))
No Yes
0.8257576 0.1742424 Weather data
tmax <- open_dataset(sources = "../weather-data/parquet/brdwgd/tmax.parquet") %>%
filter(code_muni == 3304557) %>%
filter(name == "Tmax_mean") %>%
select(date, value) %>%
collect() %>%
filter(date >= min(dengue_rj$date) & date <= max(dengue_rj$date)) %>%
summarise_by_time(.date_var = date, .by = "month", value = mean(value, na.rm = TRUE)) %>%
rename(tmax = value)
prec <- open_dataset(sources = "../weather-data/parquet/brdwgd/pr.parquet") %>%
filter(code_muni == 3304557) %>%
filter(name == "pr_sum") %>%
select(date, value) %>%
collect() %>%
filter(date >= min(dengue_rj$date) & date <= max(dengue_rj$date)) %>%
summarise_by_time(.date_var = date, .by = "month", value = sum(value, na.rm = TRUE)) %>%
rename(prec = value)
prec_avg <- open_dataset(sources = "../weather-data/parquet/brdwgd/pr.parquet") %>%
filter(code_muni == 3304557) %>%
filter(name == "pr_mean") %>%
select(date, value) %>%
collect() %>%
filter(date >= min(dengue_rj$date) & date <= max(dengue_rj$date)) %>%
summarise_by_time(.date_var = date, .by = "month", value = mean(value, na.rm = TRUE)) %>%
rename(prec_avg = value)
rh <- open_dataset(sources = "../weather-data/parquet/brdwgd/rh.parquet") %>%
filter(code_muni == 3304557) %>%
filter(name == "RH_mean") %>%
select(date, value) %>%
collect() %>%
filter(date >= min(dengue_rj$date) & date <= max(dengue_rj$date)) %>%
summarise_by_time(.date_var = date, .by = "month", value = mean(value, na.rm = TRUE)) %>%
rename(rh = value)
wind <- open_dataset(sources = "../weather-data/parquet/brdwgd/u2.parquet") %>%
filter(code_muni == 3304557) %>%
filter(name == "u2_mean") %>%
select(date, value) %>%
collect() %>%
filter(date >= min(dengue_rj$date) & date <= max(dengue_rj$date)) %>%
summarise_by_time(.date_var = date, .by = "month", value = mean(value, na.rm = TRUE)) %>%
rename(wind = value)plot_time_series(.data = tmax, .date_var = date, .value = tmax, .smooth = FALSE, .title = "Max temp, average")plot_time_series(.data = prec, .date_var = date, .value = prec, .smooth = FALSE, .title = "Precipitation, sum")plot_time_series(.data = prec_avg, .date_var = date, .value = prec_avg, .smooth = FALSE, .title = "Precipitation, average")plot_time_series(.data = rh, .date_var = date, .value = rh, .smooth = FALSE, .title = "Relative humidity, average")plot_time_series(.data = wind, .date_var = date, .value = wind, .smooth = FALSE, .title = "Wind, average")Join data
res <- inner_join(x = dengue_rj, y = tmax, by = "date") %>%
inner_join(prec, by = "date") %>%
inner_join(prec_avg, by = "date") %>%
inner_join(rh, by = "date") %>%
inner_join(wind, by = "date") %>%
select(date, anomaly, tmax, prec, rh, wind)Decision tree
Prepare data
Remove date
Lag variables: 12 months
res_prep <- res %>%
select(-date) %>%
tk_augment_lags(.value = c(tmax, prec, rh, wind), .lags = 1:12)Parameters
tree_spec <- decision_tree() %>%
set_engine("rpart") %>%
set_mode("classification")Fit model
fit1 <- tree_spec %>%
fit(anomaly ~ ., data = res_prep, model = TRUE)fit1 %>% extract_fit_engine() %>%
rpart.plot(roundint = FALSE)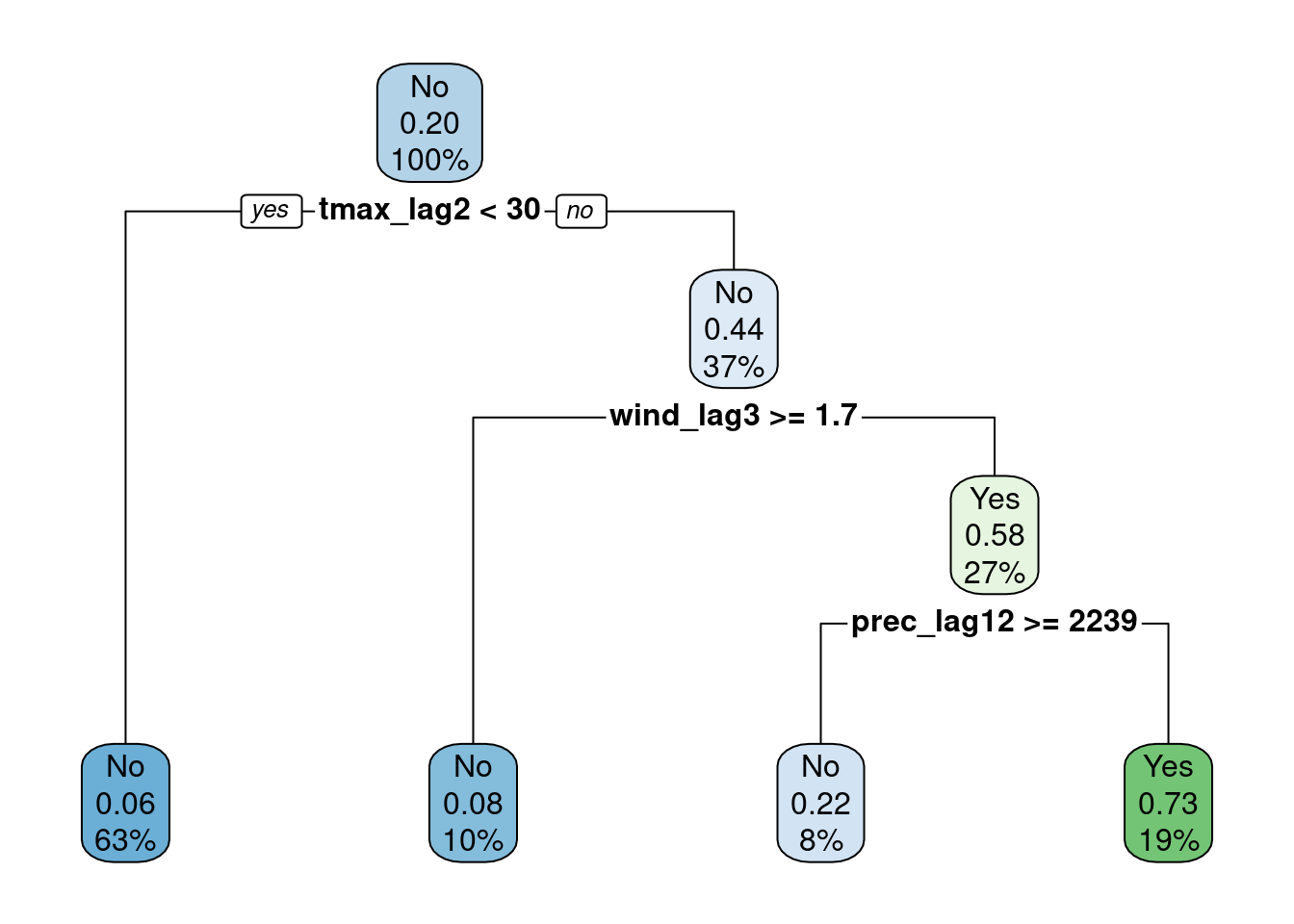
augment(fit1, new_data = res_prep) %>%
conf_mat(truth = anomaly, estimate = .pred_class) Truth
Prediction No Yes
No 86 7
Yes 6 16augment(fit1, new_data = res_prep) %>%
accuracy(truth = anomaly, estimate = .pred_class)# A tibble: 1 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 accuracy binary 0.887fit1 %>%
extract_fit_engine() %>%
vip()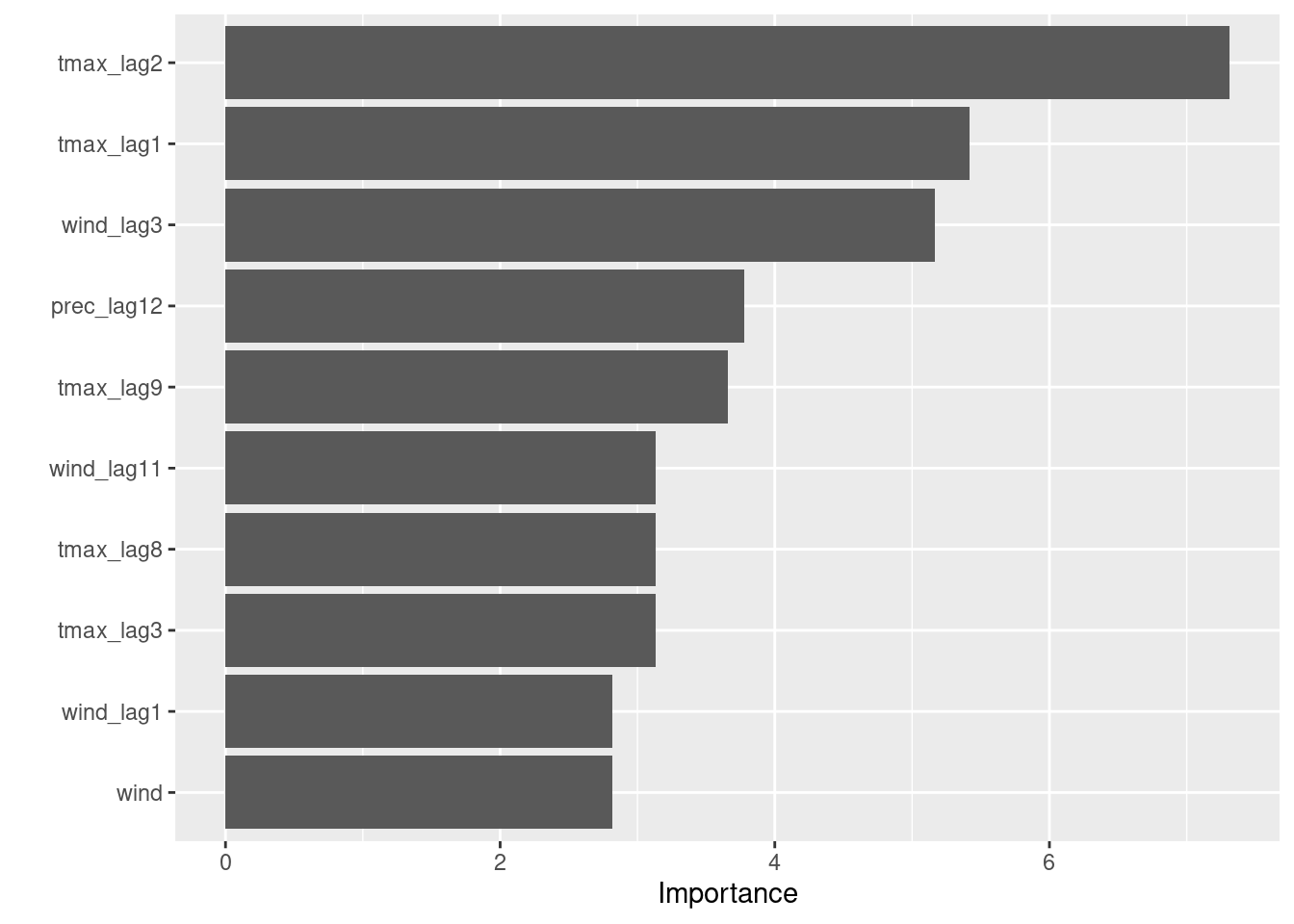
Session info
sessionInfo()R version 4.1.2 (2021-11-01)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 22.04.2 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.10.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.10.0
locale:
[1] LC_CTYPE=pt_BR.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] vip_0.3.2 rpart.plot_3.1.1 rpart_4.1.16 timetk_2.8.3
[5] arrow_12.0.1 yardstick_1.2.0 workflowsets_1.0.1 workflows_1.1.3
[9] tune_1.1.1 rsample_1.1.1 recipes_1.0.6 parsnip_1.1.0
[13] modeldata_1.1.0 infer_1.0.4 dials_1.2.0 scales_1.2.1
[17] broom_1.0.5 tidymodels_1.1.0 lubridate_1.9.2 forcats_1.0.0
[21] stringr_1.5.0 dplyr_1.1.2 purrr_1.0.1 readr_2.1.4
[25] tidyr_1.3.0 tibble_3.2.1 ggplot2_3.4.2 tidyverse_2.0.0
loaded via a namespace (and not attached):
[1] xts_0.13.1 bit64_4.0.5 httr_1.4.6
[4] DiceDesign_1.9 tools_4.1.2 backports_1.4.1
[7] utf8_1.2.3 R6_2.5.1 lazyeval_0.2.2
[10] colorspace_2.1-0 nnet_7.3-17 withr_2.5.0
[13] gridExtra_2.3 tidyselect_1.2.0 bit_4.0.5
[16] compiler_4.1.2 cli_3.6.1 plotly_4.10.2
[19] labeling_0.4.2 digest_0.6.32 rmarkdown_2.23
[22] pkgconfig_2.0.3 htmltools_0.5.5 parallelly_1.36.0
[25] lhs_1.1.6 fastmap_1.1.1 htmlwidgets_1.6.2
[28] rlang_1.1.1 rstudioapi_0.14 farver_2.1.1
[31] generics_0.1.3 zoo_1.8-12 jsonlite_1.8.7
[34] crosstalk_1.2.0 magrittr_2.0.3 Matrix_1.5-4.1
[37] Rcpp_1.0.10 munsell_0.5.0 fansi_1.0.4
[40] GPfit_1.0-8 lifecycle_1.0.3 furrr_0.3.1
[43] stringi_1.7.12 yaml_2.3.7 MASS_7.3-55
[46] grid_4.1.2 parallel_4.1.2 listenv_0.9.0
[49] lattice_0.20-45 splines_4.1.2 hms_1.1.3
[52] knitr_1.43 pillar_1.9.0 future.apply_1.11.0
[55] codetools_0.2-18 glue_1.6.2 evaluate_0.21
[58] data.table_1.14.8 vctrs_0.6.3 tzdb_0.4.0
[61] foreach_1.5.2 gtable_0.3.3 future_1.33.0
[64] assertthat_0.2.1 xfun_0.39 gower_1.0.1
[67] prodlim_2023.03.31 viridisLite_0.4.2 class_7.3-20
[70] survival_3.2-13 timeDate_4022.108 iterators_1.0.14
[73] hardhat_1.3.0 lava_1.7.2.1 timechange_0.2.0
[76] globals_0.16.2 ellipsis_0.3.2 ipred_0.9-14